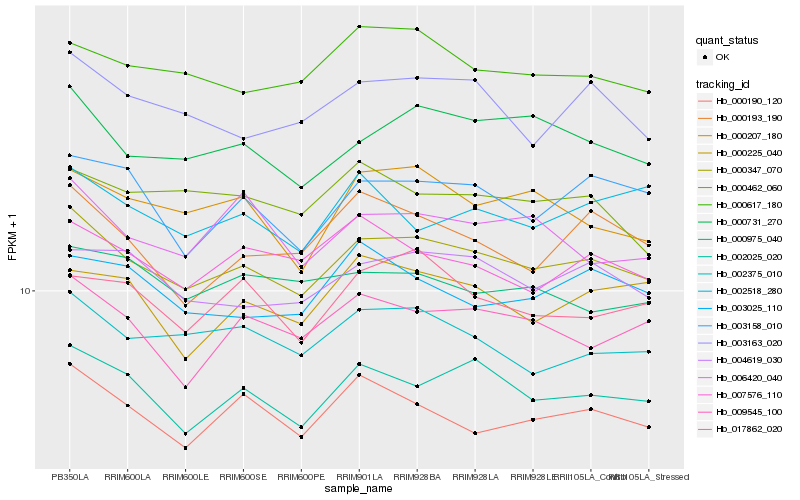
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000347_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002025_020 |
0.0495944333 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g39710 [Jatropha curcas] |
| 3 |
Hb_000731_270 |
0.0498114494 |
- |
- |
PREDICTED: polyadenylate-binding protein RBP45B isoform X1 [Jatropha curcas] |
| 4 |
Hb_004619_030 |
0.0547319132 |
- |
- |
PREDICTED: uncharacterized protein LOC105633364 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002375_010 |
0.0555551501 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003163_020 |
0.0560135408 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 12-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000190_120 |
0.0580421094 |
- |
- |
PREDICTED: uncharacterized protein LOC105649936 [Jatropha curcas] |
| 8 |
Hb_003158_010 |
0.0581616777 |
- |
- |
hypothetical protein JCGZ_17361 [Jatropha curcas] |
| 9 |
Hb_000617_180 |
0.0589255665 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 10 |
Hb_009545_100 |
0.0594194302 |
- |
- |
PREDICTED: protein OBERON 1 [Jatropha curcas] |
| 11 |
Hb_000207_180 |
0.0605592088 |
- |
- |
PREDICTED: calponin homology domain-containing protein DDB_G0272472 [Jatropha curcas] |
| 12 |
Hb_007576_110 |
0.0607938586 |
- |
- |
Endoplasmic reticulum vesicle transporter protein [Theobroma cacao] |
| 13 |
Hb_000193_190 |
0.0639467765 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 16 [Jatropha curcas] |
| 14 |
Hb_006420_040 |
0.0645600031 |
transcription factor |
TF Family: Orphans |
PREDICTED: paired amphipathic helix protein Sin3-like 4 isoform X2 [Jatropha curcas] |
| 15 |
Hb_017862_020 |
0.0660594411 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase lvsG [Jatropha curcas] |
| 16 |
Hb_003025_110 |
0.0663687082 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000462_060 |
0.066772691 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000975_040 |
0.0672476877 |
- |
- |
hypothetical protein CICLE_v10028249mg [Citrus clementina] |
| 19 |
Hb_000225_040 |
0.0677633109 |
- |
- |
PREDICTED: translation factor GUF1 homolog, mitochondrial [Nelumbo nucifera] |
| 20 |
Hb_002518_280 |
0.0693457138 |
- |
- |
phosphate transporter [Manihot esculenta] |