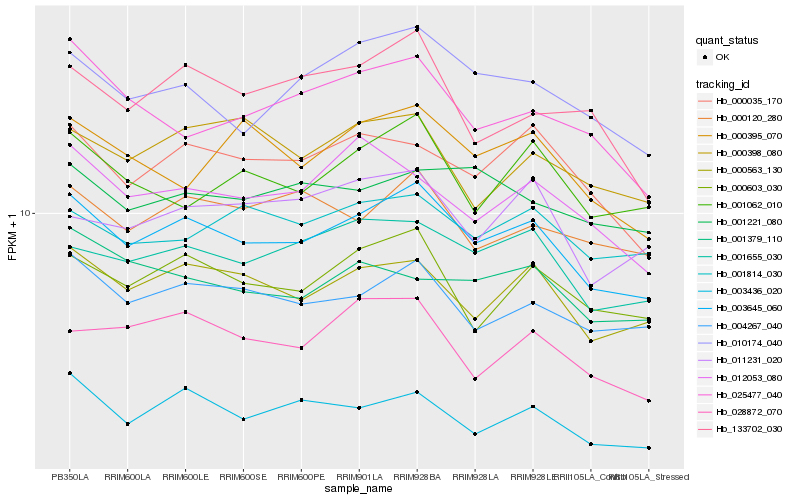
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003645_060 |
0.0 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000563_130 |
0.0574514877 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 3 |
Hb_004267_040 |
0.0613298974 |
- |
- |
PREDICTED: transcriptional adapter ADA2a [Jatropha curcas] |
| 4 |
Hb_001655_030 |
0.0616722999 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000603_030 |
0.0621072596 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 6 |
Hb_010174_040 |
0.0627837018 |
- |
- |
PREDICTED: T-complex protein 1 subunit epsilon [Jatropha curcas] |
| 7 |
Hb_000035_170 |
0.0658903422 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 2-like [Jatropha curcas] |
| 8 |
Hb_003436_020 |
0.0664883256 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105636276 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000120_280 |
0.0746990166 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 10 |
Hb_012053_080 |
0.075269313 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 11 |
Hb_001062_010 |
0.0757100829 |
- |
- |
PREDICTED: CWF19-like protein 2 [Jatropha curcas] |
| 12 |
Hb_028872_070 |
0.0763275885 |
desease resistance |
Gene Name: DEAD |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase PRP1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001814_030 |
0.0767393926 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 14 |
Hb_000395_070 |
0.0771237612 |
- |
- |
PREDICTED: serine/threonine-protein kinase BLUS1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_025477_040 |
0.0790184125 |
- |
- |
PREDICTED: post-GPI attachment to proteins factor 3 [Jatropha curcas] |
| 16 |
Hb_001221_080 |
0.079534624 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_133702_030 |
0.0795932301 |
- |
- |
PREDICTED: uric acid degradation bifunctional protein TTL isoform X1 [Jatropha curcas] |
| 18 |
Hb_000398_080 |
0.0800233894 |
- |
- |
tip120, putative [Ricinus communis] |
| 19 |
Hb_011231_020 |
0.0802579173 |
- |
- |
PREDICTED: proline-, glutamic acid- and leucine-rich protein 1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001379_110 |
0.0819998363 |
- |
- |
PREDICTED: sec1 family domain-containing protein MIP3 [Jatropha curcas] |