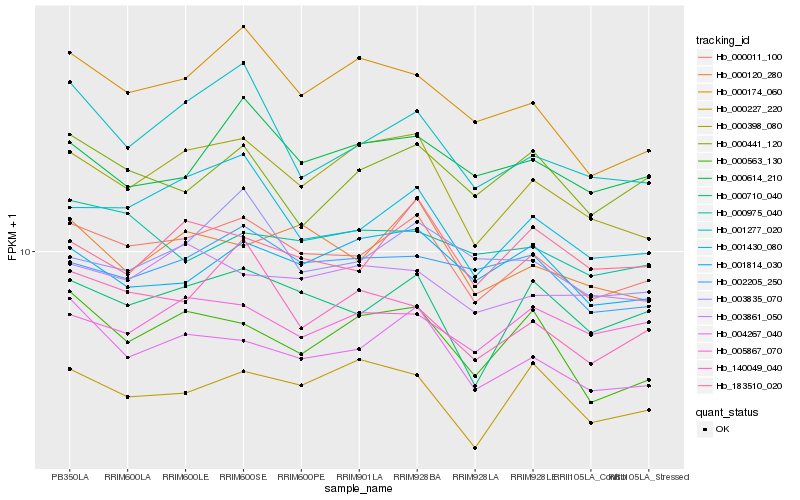
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000011_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 2 |
Hb_004267_040 |
0.0446715111 |
- |
- |
PREDICTED: transcriptional adapter ADA2a [Jatropha curcas] |
| 3 |
Hb_000710_040 |
0.0472385125 |
- |
- |
PREDICTED: HEAT repeat-containing protein 6 [Jatropha curcas] |
| 4 |
Hb_001430_080 |
0.0497750068 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 5 |
Hb_000398_080 |
0.0502174685 |
- |
- |
tip120, putative [Ricinus communis] |
| 6 |
Hb_000120_280 |
0.0561589918 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 7 |
Hb_000563_130 |
0.0607722218 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 8 |
Hb_002205_250 |
0.0614915566 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_183510_020 |
0.0616784267 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 10 |
Hb_003861_050 |
0.0623392099 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 11 |
Hb_000174_060 |
0.063067414 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 12 |
Hb_000227_220 |
0.0632644964 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 13 |
Hb_001814_030 |
0.0635400277 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 14 |
Hb_000614_210 |
0.0649732812 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_140049_040 |
0.0652418176 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 16 |
Hb_005867_070 |
0.0660321725 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_001277_020 |
0.0660927169 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |
| 18 |
Hb_000975_040 |
0.0662993833 |
- |
- |
hypothetical protein CICLE_v10028249mg [Citrus clementina] |
| 19 |
Hb_003835_070 |
0.0670724907 |
- |
- |
PREDICTED: uncharacterized protein LOC105641698 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000441_120 |
0.0678353591 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT111, chloroplastic isoform X1 [Jatropha curcas] |