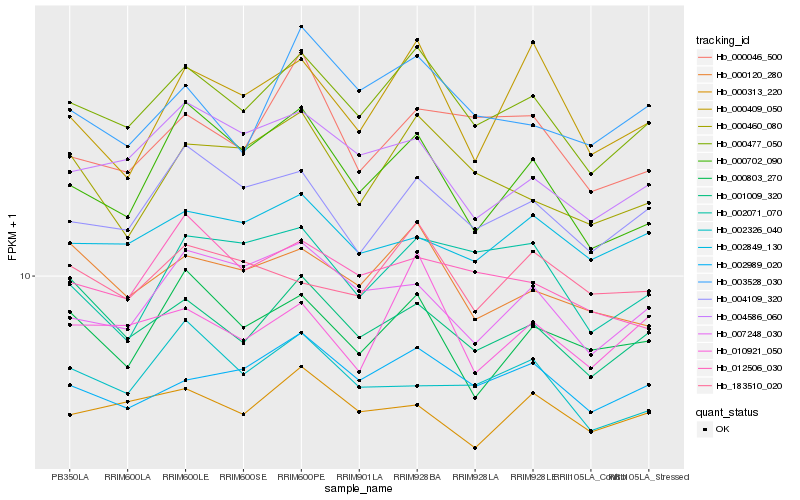
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000477_050 |
0.0 |
- |
- |
PREDICTED: 26S protease regulatory subunit 8 homolog A [Jatropha curcas] |
| 2 |
Hb_002989_020 |
0.0536879945 |
- |
- |
PREDICTED: uncharacterized protein LOC105630013 isoform X2 [Jatropha curcas] |
| 3 |
Hb_004109_320 |
0.059228692 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 4 |
Hb_001009_320 |
0.0599131987 |
- |
- |
PREDICTED: uncharacterized protein LOC105640250 [Jatropha curcas] |
| 5 |
Hb_000120_280 |
0.060442947 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 6 |
Hb_000460_080 |
0.0626733162 |
- |
- |
acyl-coenzyme A binding domain containing, putative [Ricinus communis] |
| 7 |
Hb_003528_030 |
0.0653616191 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit L-like [Jatropha curcas] |
| 8 |
Hb_010921_050 |
0.0658177258 |
- |
- |
PREDICTED: protein POLLEN DEFECTIVE IN GUIDANCE 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000702_090 |
0.0659677489 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 10 |
Hb_007248_030 |
0.0669648473 |
- |
- |
PREDICTED: uncharacterized protein LOC105633558 [Jatropha curcas] |
| 11 |
Hb_012506_030 |
0.0673572938 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 12 |
Hb_004586_060 |
0.0674681131 |
- |
- |
PREDICTED: AP-4 complex subunit mu [Jatropha curcas] |
| 13 |
Hb_000046_500 |
0.0688770174 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Jatropha curcas] |
| 14 |
Hb_000803_270 |
0.0700518426 |
- |
- |
PREDICTED: nuclear cap-binding protein subunit 1 [Jatropha curcas] |
| 15 |
Hb_000313_220 |
0.0704702482 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002071_070 |
0.0708395126 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_183510_020 |
0.0720812698 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 18 |
Hb_002849_130 |
0.0723387121 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 19 |
Hb_002326_040 |
0.073075586 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 20 |
Hb_000409_050 |
0.0732757778 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |