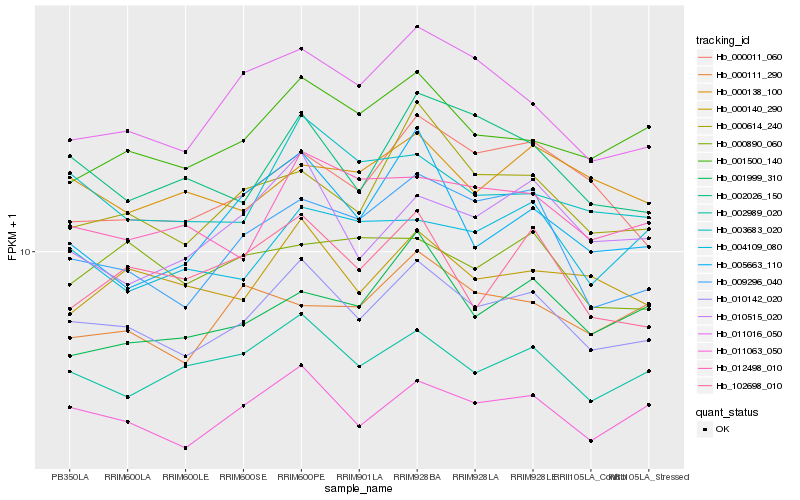
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010142_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_011063_050 |
0.0392655736 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17670 [Jatropha curcas] |
| 3 |
Hb_002989_020 |
0.0503158482 |
- |
- |
PREDICTED: uncharacterized protein LOC105630013 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000614_240 |
0.0547756097 |
- |
- |
PREDICTED: signal recognition particle subunit SRP72 [Jatropha curcas] |
| 5 |
Hb_001500_140 |
0.0548177016 |
- |
- |
pelota, putative [Ricinus communis] |
| 6 |
Hb_000140_290 |
0.0567465926 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 59 kDa protein isoform X1 [Populus euphratica] |
| 7 |
Hb_003683_020 |
0.0571341988 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 8 |
Hb_002026_150 |
0.0593171392 |
transcription factor |
TF Family: SNF2 |
PREDICTED: DNA repair helicase XPB1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_009296_040 |
0.0594386908 |
- |
- |
hypothetical protein L484_007435 [Morus notabilis] |
| 10 |
Hb_012498_010 |
0.0596441844 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM44-2 [Jatropha curcas] |
| 11 |
Hb_011016_050 |
0.0611487319 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 isoform X1 [Jatropha curcas] |
| 12 |
Hb_010515_020 |
0.0618980464 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 13 |
Hb_000011_060 |
0.0621875466 |
- |
- |
PREDICTED: exportin-7 isoform X1 [Jatropha curcas] |
| 14 |
Hb_102698_010 |
0.0657529785 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000138_100 |
0.0664974608 |
- |
- |
bifunctional purine biosynthesis protein, putative [Ricinus communis] |
| 16 |
Hb_001999_310 |
0.0665881337 |
- |
- |
PREDICTED: lysosomal beta glucosidase-like [Jatropha curcas] |
| 17 |
Hb_004109_080 |
0.0689160228 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 15-like isoform X1 [Populus euphratica] |
| 18 |
Hb_000890_060 |
0.0690847881 |
- |
- |
PREDICTED: sucrose nonfermenting 4-like protein [Jatropha curcas] |
| 19 |
Hb_000111_290 |
0.0693795183 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase KEG [Jatropha curcas] |
| 20 |
Hb_005663_110 |
0.069521046 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X2 [Jatropha curcas] |