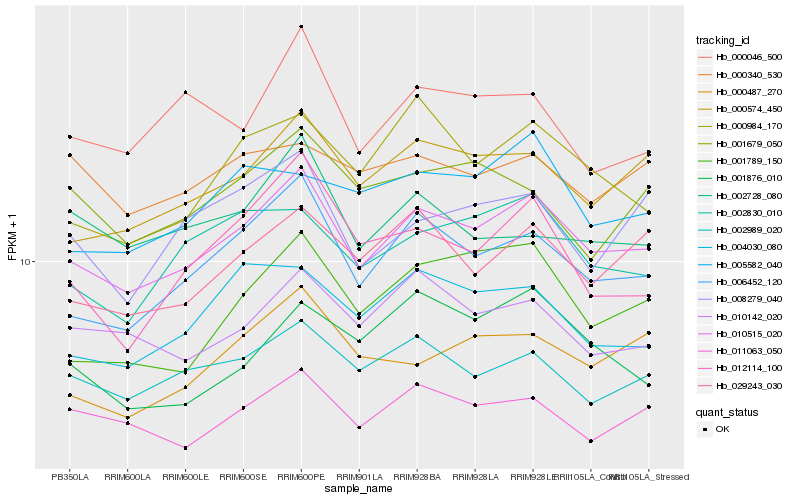
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010515_020 |
0.0 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 2 |
Hb_006452_120 |
0.0457268037 |
- |
- |
PREDICTED: protein FAM188A [Jatropha curcas] |
| 3 |
Hb_000574_450 |
0.0541928327 |
- |
- |
PREDICTED: signal recognition particle 54 kDa protein 2 [Jatropha curcas] |
| 4 |
Hb_002989_020 |
0.0561248635 |
- |
- |
PREDICTED: uncharacterized protein LOC105630013 isoform X2 [Jatropha curcas] |
| 5 |
Hb_011063_050 |
0.0586975637 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17670 [Jatropha curcas] |
| 6 |
Hb_029243_030 |
0.0602549436 |
- |
- |
PREDICTED: regulatory-associated protein of TOR 1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002830_010 |
0.0617120605 |
- |
- |
PREDICTED: TBCC domain-containing protein 1 [Jatropha curcas] |
| 8 |
Hb_010142_020 |
0.0618980464 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000487_270 |
0.06296201 |
- |
- |
hypothetical protein JCGZ_22651 [Jatropha curcas] |
| 10 |
Hb_004030_080 |
0.0638305231 |
- |
- |
hypothetical protein JCGZ_25110 [Jatropha curcas] |
| 11 |
Hb_000984_170 |
0.0659191001 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 12 |
Hb_001789_150 |
0.0667345305 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_005582_040 |
0.0678423676 |
- |
- |
PREDICTED: regulator of nonsense transcripts 1 homolog [Jatropha curcas] |
| 14 |
Hb_008279_040 |
0.0683970724 |
- |
- |
Rer1 family protein isoform 2 [Theobroma cacao] |
| 15 |
Hb_000046_500 |
0.0706008853 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Jatropha curcas] |
| 16 |
Hb_012114_100 |
0.0715568167 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 17 |
Hb_000340_530 |
0.0724257684 |
- |
- |
hypothetical protein VITISV_016664 [Vitis vinifera] |
| 18 |
Hb_001679_050 |
0.072838152 |
- |
- |
PREDICTED: exocyst complex component EXO84B [Jatropha curcas] |
| 19 |
Hb_002728_080 |
0.0738392403 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 20 |
Hb_001876_010 |
0.0748146029 |
- |
- |
PREDICTED: arginyl-tRNA--protein transferase 2-like isoform X1 [Jatropha curcas] |