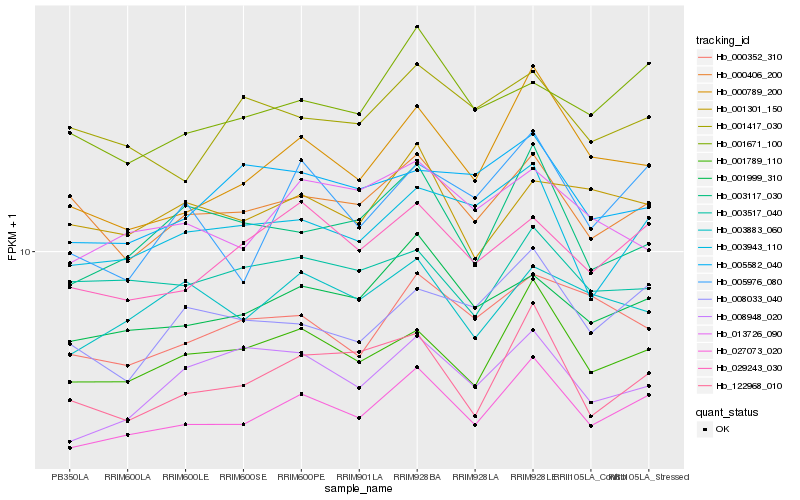
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027073_020 |
0.0 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 2 |
Hb_001789_110 |
0.0521859531 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_001999_310 |
0.0531590189 |
- |
- |
PREDICTED: lysosomal beta glucosidase-like [Jatropha curcas] |
| 4 |
Hb_000789_200 |
0.0603336207 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 5 |
Hb_029243_030 |
0.0613725193 |
- |
- |
PREDICTED: regulatory-associated protein of TOR 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_122968_010 |
0.0643865918 |
- |
- |
hypothetical protein JCGZ_23576 [Jatropha curcas] |
| 7 |
Hb_000406_200 |
0.0660791277 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit epsilon-like isoform X4 [Jatropha curcas] |
| 8 |
Hb_003943_110 |
0.0671190077 |
transcription factor |
TF Family: MYB-related |
Zuotin, putative [Ricinus communis] |
| 9 |
Hb_001671_100 |
0.0696308693 |
- |
- |
glycine-rich RNA-binding family protein [Populus trichocarpa] |
| 10 |
Hb_003883_060 |
0.0701850633 |
transcription factor |
TF Family: BSD |
PREDICTED: probable RNA polymerase II transcription factor B subunit 1-1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_008033_040 |
0.0707222745 |
- |
- |
PREDICTED: MATE efflux family protein 3, chloroplastic [Jatropha curcas] |
| 12 |
Hb_003117_030 |
0.0718500362 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 13 |
Hb_000352_310 |
0.0722508397 |
- |
- |
hypothetical protein CISIN_1g0017891mg, partial [Citrus sinensis] |
| 14 |
Hb_013726_090 |
0.0746739962 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 15 |
Hb_008948_020 |
0.0747123073 |
- |
- |
hypothetical protein JCGZ_21216 [Jatropha curcas] |
| 16 |
Hb_003517_040 |
0.0753699453 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 17 |
Hb_005976_080 |
0.0758922005 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 18 |
Hb_001301_150 |
0.0763495112 |
- |
- |
PREDICTED: UPF0136 membrane protein At2g26240 [Vitis vinifera] |
| 19 |
Hb_005582_040 |
0.0773945717 |
- |
- |
PREDICTED: regulator of nonsense transcripts 1 homolog [Jatropha curcas] |
| 20 |
Hb_001417_030 |
0.0774229182 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |