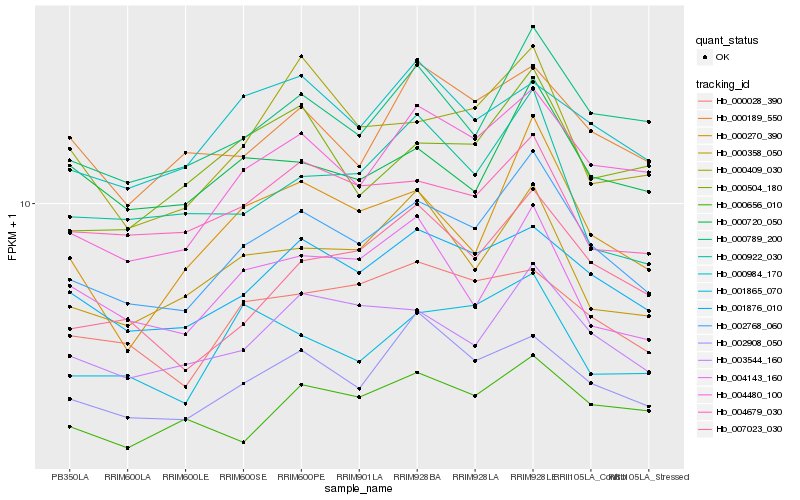
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002768_060 |
0.0 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001876_010 |
0.0637067632 |
- |
- |
PREDICTED: arginyl-tRNA--protein transferase 2-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_004480_100 |
0.0659099658 |
- |
- |
PREDICTED: calcium homeostasis endoplasmic reticulum protein [Jatropha curcas] |
| 4 |
Hb_003544_160 |
0.0659297902 |
- |
- |
- |
| 5 |
Hb_000789_200 |
0.066243322 |
- |
- |
mitochondrial processing peptidase alpha subunit, putative [Ricinus communis] |
| 6 |
Hb_002908_050 |
0.075739368 |
- |
- |
hypothetical protein CISIN_1g0022902mg, partial [Citrus sinensis] |
| 7 |
Hb_004679_030 |
0.0782511442 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 8 |
Hb_000358_050 |
0.0783418164 |
- |
- |
PREDICTED: uncharacterized protein LOC105633378 [Jatropha curcas] |
| 9 |
Hb_000270_390 |
0.0799620391 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 10 |
Hb_000028_390 |
0.0815042315 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 3-like [Jatropha curcas] |
| 11 |
Hb_000720_050 |
0.0834042533 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000409_030 |
0.0839976289 |
- |
- |
PREDICTED: uncharacterized protein LOC105642693 [Jatropha curcas] |
| 13 |
Hb_007023_030 |
0.0840632258 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g20710, mitochondrial-like [Jatropha curcas] |
| 14 |
Hb_001865_070 |
0.0853212695 |
- |
- |
PREDICTED: uncharacterized protein LOC105647937 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000189_550 |
0.0860302025 |
- |
- |
PREDICTED: protein transport protein Sec24-like At4g32640 [Jatropha curcas] |
| 16 |
Hb_000984_170 |
0.0860376304 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 17 |
Hb_004143_160 |
0.0866436784 |
- |
- |
SAB, putative [Ricinus communis] |
| 18 |
Hb_000504_180 |
0.0868079437 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 19 |
Hb_000656_010 |
0.0877561519 |
- |
- |
PREDICTED: uncharacterized protein LOC105631354 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000922_030 |
0.0896583887 |
- |
- |
PREDICTED: protein EXECUTER 2, chloroplastic [Jatropha curcas] |