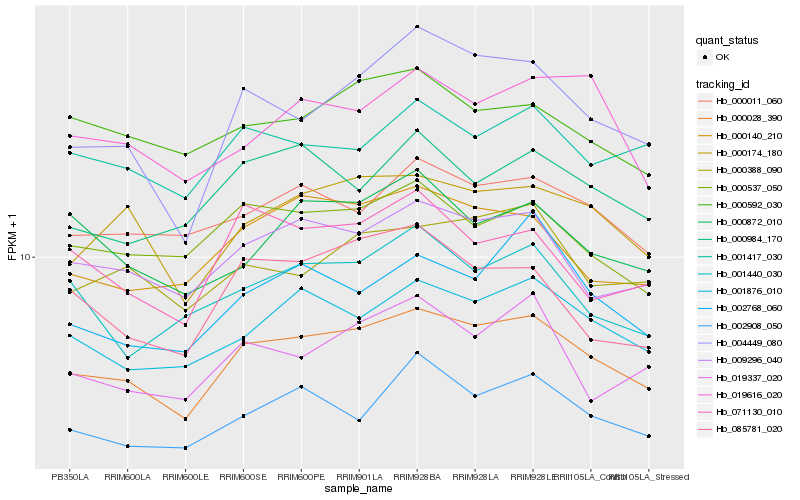
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000028_390 |
0.0 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 3-like [Jatropha curcas] |
| 2 |
Hb_009296_040 |
0.0615995606 |
- |
- |
hypothetical protein L484_007435 [Morus notabilis] |
| 3 |
Hb_001876_010 |
0.0626473423 |
- |
- |
PREDICTED: arginyl-tRNA--protein transferase 2-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000174_180 |
0.063914836 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_004449_080 |
0.0670071368 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH3-like [Jatropha curcas] |
| 6 |
Hb_000140_210 |
0.0684124217 |
- |
- |
sec10, putative [Ricinus communis] |
| 7 |
Hb_000011_060 |
0.0704350215 |
- |
- |
PREDICTED: exportin-7 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000388_090 |
0.0733636157 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 9 |
Hb_000537_050 |
0.0737795129 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 10 |
Hb_002908_050 |
0.0740880689 |
- |
- |
hypothetical protein CISIN_1g0022902mg, partial [Citrus sinensis] |
| 11 |
Hb_000984_170 |
0.0783380475 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 12 |
Hb_000592_030 |
0.0795630479 |
- |
- |
PREDICTED: uncharacterized protein LOC100853969 isoform X1 [Vitis vinifera] |
| 13 |
Hb_001417_030 |
0.0803101683 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 14 |
Hb_001440_030 |
0.0809017799 |
- |
- |
PREDICTED: exocyst complex component SEC8 [Jatropha curcas] |
| 15 |
Hb_002768_060 |
0.0815042315 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000872_010 |
0.0819387404 |
- |
- |
PREDICTED: putative ubiquitin-conjugating enzyme E2 38 [Jatropha curcas] |
| 17 |
Hb_019337_020 |
0.0820560023 |
- |
- |
K+ uptake permease 11 isoform 1 [Theobroma cacao] |
| 18 |
Hb_019616_020 |
0.0821513576 |
- |
- |
PREDICTED: alanine--tRNA ligase [Jatropha curcas] |
| 19 |
Hb_085781_020 |
0.0823799837 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1 [Jatropha curcas] |
| 20 |
Hb_071130_010 |
0.0832617882 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |