| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
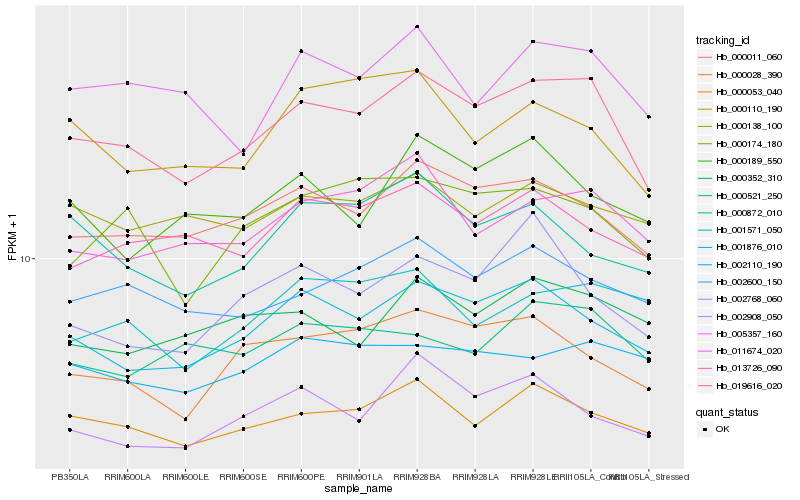
Hb_019616_020 |
0.0 |
- |
- |
PREDICTED: alanine--tRNA ligase [Jatropha curcas] |
| 2 |
Hb_000011_060 |
0.0672320132 |
- |
- |
PREDICTED: exportin-7 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001876_010 |
0.069542885 |
- |
- |
PREDICTED: arginyl-tRNA--protein transferase 2-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000053_040 |
0.0728105069 |
- |
- |
hypothetical protein CISIN_1g0010392mg, partial [Citrus sinensis] |
| 5 |
Hb_011674_020 |
0.0808643732 |
- |
- |
PREDICTED: NEDD8-activating enzyme E1 regulatory subunit-like [Jatropha curcas] |
| 6 |
Hb_002600_150 |
0.081359576 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000028_390 |
0.0821513576 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 3-like [Jatropha curcas] |
| 8 |
Hb_000174_180 |
0.0847002998 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_005357_160 |
0.0847812208 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 10 |
Hb_013726_090 |
0.0875812272 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase At1g63170 [Jatropha curcas] |
| 11 |
Hb_002908_050 |
0.0891669093 |
- |
- |
hypothetical protein CISIN_1g0022902mg, partial [Citrus sinensis] |
| 12 |
Hb_000110_190 |
0.0926102406 |
- |
- |
hypothetical protein B456_013G081300 [Gossypium raimondii] |
| 13 |
Hb_000138_100 |
0.0943078215 |
- |
- |
bifunctional purine biosynthesis protein, putative [Ricinus communis] |
| 14 |
Hb_000352_310 |
0.0944230113 |
- |
- |
hypothetical protein CISIN_1g0017891mg, partial [Citrus sinensis] |
| 15 |
Hb_000872_010 |
0.0944621666 |
- |
- |
PREDICTED: putative ubiquitin-conjugating enzyme E2 38 [Jatropha curcas] |
| 16 |
Hb_000521_250 |
0.0954818582 |
- |
- |
PREDICTED: protein N-terminal asparagine amidohydrolase isoform X2 [Jatropha curcas] |
| 17 |
Hb_002110_190 |
0.0969551768 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27460 [Jatropha curcas] |
| 18 |
Hb_000189_550 |
0.0971513304 |
- |
- |
PREDICTED: protein transport protein Sec24-like At4g32640 [Jatropha curcas] |
| 19 |
Hb_001571_050 |
0.0978278078 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3-like [Populus euphratica] |
| 20 |
Hb_002768_060 |
0.0978572297 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 2 isoform X1 [Jatropha curcas] |