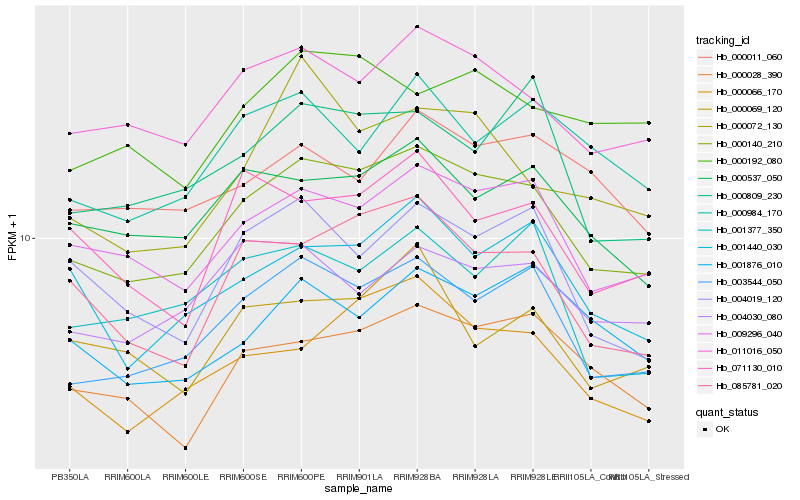
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000140_210 |
0.0 |
- |
- |
sec10, putative [Ricinus communis] |
| 2 |
Hb_003544_050 |
0.0452924615 |
- |
- |
PREDICTED: uncharacterized protein LOC105644300 isoform X1 [Jatropha curcas] |
| 3 |
Hb_009296_040 |
0.0529531677 |
- |
- |
hypothetical protein L484_007435 [Morus notabilis] |
| 4 |
Hb_011016_050 |
0.0560957757 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000066_170 |
0.0633223579 |
- |
- |
PREDICTED: phosphatidylinositol-3-phosphatase myotubularin-1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_085781_020 |
0.0638534914 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1 [Jatropha curcas] |
| 7 |
Hb_000028_390 |
0.0684124217 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 3-like [Jatropha curcas] |
| 8 |
Hb_000537_050 |
0.0697726882 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 9 |
Hb_001440_030 |
0.0713240237 |
- |
- |
PREDICTED: exocyst complex component SEC8 [Jatropha curcas] |
| 10 |
Hb_000809_230 |
0.0747806766 |
- |
- |
PREDICTED: uncharacterized protein LOC105639681 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004019_120 |
0.0753983278 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |
| 12 |
Hb_004030_080 |
0.0754323932 |
- |
- |
hypothetical protein JCGZ_25110 [Jatropha curcas] |
| 13 |
Hb_000984_170 |
0.0770880086 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 14 |
Hb_071130_010 |
0.0785516068 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000072_130 |
0.0802302041 |
- |
- |
hypothetical protein OsJ_28567 [Oryza sativa Japonica Group] |
| 16 |
Hb_000069_120 |
0.0803218605 |
- |
- |
PREDICTED: uncharacterized protein LOC105124742 isoform X1 [Populus euphratica] |
| 17 |
Hb_001377_350 |
0.0805639394 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 18 |
Hb_001876_010 |
0.0810768322 |
- |
- |
PREDICTED: arginyl-tRNA--protein transferase 2-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000192_080 |
0.0823075565 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK3 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000011_060 |
0.0824678151 |
- |
- |
PREDICTED: exportin-7 isoform X1 [Jatropha curcas] |