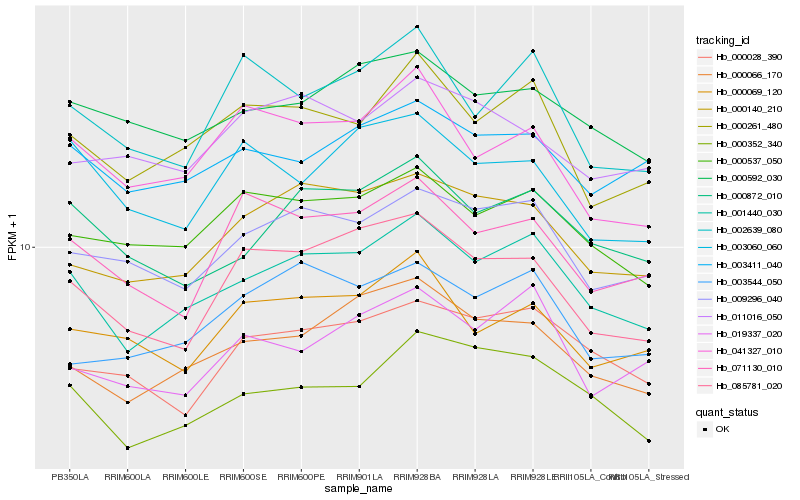
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000066_170 |
0.0 |
- |
- |
PREDICTED: phosphatidylinositol-3-phosphatase myotubularin-1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001440_030 |
0.0624034174 |
- |
- |
PREDICTED: exocyst complex component SEC8 [Jatropha curcas] |
| 3 |
Hb_000140_210 |
0.0633223579 |
- |
- |
sec10, putative [Ricinus communis] |
| 4 |
Hb_085781_020 |
0.0674078355 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 1 [Jatropha curcas] |
| 5 |
Hb_000592_030 |
0.07727606 |
- |
- |
PREDICTED: uncharacterized protein LOC100853969 isoform X1 [Vitis vinifera] |
| 6 |
Hb_000537_050 |
0.0794996605 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 7 |
Hb_009296_040 |
0.0861994838 |
- |
- |
hypothetical protein L484_007435 [Morus notabilis] |
| 8 |
Hb_000028_390 |
0.0870836084 |
- |
- |
PREDICTED: extra-large guanine nucleotide-binding protein 3-like [Jatropha curcas] |
| 9 |
Hb_003544_050 |
0.0874242627 |
- |
- |
PREDICTED: uncharacterized protein LOC105644300 isoform X1 [Jatropha curcas] |
| 10 |
Hb_003411_040 |
0.0875271272 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_019337_020 |
0.0883014059 |
- |
- |
K+ uptake permease 11 isoform 1 [Theobroma cacao] |
| 12 |
Hb_000261_480 |
0.0883602974 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 13 |
Hb_000352_340 |
0.0887982152 |
- |
- |
PREDICTED: importin-13 [Jatropha curcas] |
| 14 |
Hb_000069_120 |
0.0907935273 |
- |
- |
PREDICTED: uncharacterized protein LOC105124742 isoform X1 [Populus euphratica] |
| 15 |
Hb_041327_010 |
0.0919387405 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 16 |
Hb_011016_050 |
0.0939035199 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000872_010 |
0.0943608526 |
- |
- |
PREDICTED: putative ubiquitin-conjugating enzyme E2 38 [Jatropha curcas] |
| 18 |
Hb_002639_080 |
0.0948240582 |
- |
- |
PREDICTED: large proline-rich protein BAG6 isoform X1 [Jatropha curcas] |
| 19 |
Hb_071130_010 |
0.0950228106 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003060_060 |
0.0954594679 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |