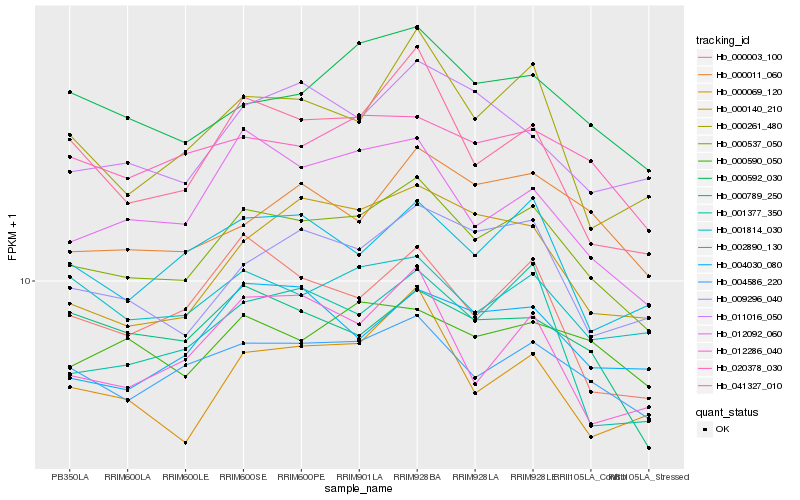
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000537_050 |
0.0 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 2 |
Hb_004586_220 |
0.0464386167 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 3 |
Hb_000592_030 |
0.0559860902 |
- |
- |
PREDICTED: uncharacterized protein LOC100853969 isoform X1 [Vitis vinifera] |
| 4 |
Hb_020378_030 |
0.0571476212 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 5 |
Hb_012092_060 |
0.0598848986 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 6 |
Hb_009296_040 |
0.0618094499 |
- |
- |
hypothetical protein L484_007435 [Morus notabilis] |
| 7 |
Hb_002890_130 |
0.0632663202 |
- |
- |
tip120, putative [Ricinus communis] |
| 8 |
Hb_000590_050 |
0.0642732465 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 9 |
Hb_000069_120 |
0.0649355013 |
- |
- |
PREDICTED: uncharacterized protein LOC105124742 isoform X1 [Populus euphratica] |
| 10 |
Hb_001814_030 |
0.0649632782 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 11 |
Hb_041327_010 |
0.0654627145 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 12 |
Hb_000011_060 |
0.0655342711 |
- |
- |
PREDICTED: exportin-7 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000003_100 |
0.0662632147 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 14 |
Hb_001377_350 |
0.0667366177 |
- |
- |
PREDICTED: exportin-2 [Jatropha curcas] |
| 15 |
Hb_004030_080 |
0.0670651404 |
- |
- |
hypothetical protein JCGZ_25110 [Jatropha curcas] |
| 16 |
Hb_011016_050 |
0.0671901013 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI7 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000261_480 |
0.0673702616 |
- |
- |
26S proteasome regulatory subunit family protein [Populus trichocarpa] |
| 18 |
Hb_012286_040 |
0.0687075403 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 19 |
Hb_000140_210 |
0.0697726882 |
- |
- |
sec10, putative [Ricinus communis] |
| 20 |
Hb_000789_250 |
0.0699545509 |
- |
- |
PREDICTED: uncharacterized protein LOC105637809 [Jatropha curcas] |