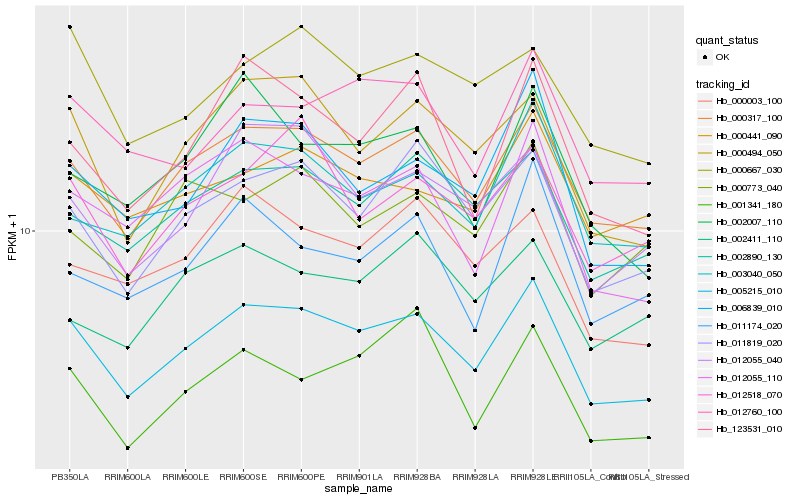
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005215_010 |
0.0 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000317_100 |
0.0271560906 |
- |
- |
PREDICTED: uncharacterized protein LOC105646995 [Jatropha curcas] |
| 3 |
Hb_002890_130 |
0.0530052459 |
- |
- |
tip120, putative [Ricinus communis] |
| 4 |
Hb_011819_020 |
0.0557661871 |
- |
- |
PREDICTED: uncharacterized protein LOC105643703 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000494_050 |
0.059614425 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 6 |
Hb_003040_050 |
0.0598627889 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 7 |
Hb_012760_100 |
0.061122011 |
- |
- |
Pre-rRNA-processing protein ESF1, putative [Ricinus communis] |
| 8 |
Hb_000441_090 |
0.0613346213 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 9 |
Hb_006839_010 |
0.061423464 |
- |
- |
PREDICTED: importin subunit beta-1-like [Jatropha curcas] |
| 10 |
Hb_012055_110 |
0.0630400165 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 11 |
Hb_012518_070 |
0.0641900988 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 9 isoform X1 [Jatropha curcas] |
| 12 |
Hb_012055_040 |
0.0651427615 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 13 |
Hb_000667_030 |
0.0652132959 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 14 |
Hb_000003_100 |
0.0676700911 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 15 |
Hb_123531_010 |
0.0703084593 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 16 |
Hb_000773_040 |
0.0706647124 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001341_180 |
0.071702213 |
- |
- |
PREDICTED: transmembrane protein 209 [Jatropha curcas] |
| 18 |
Hb_002007_110 |
0.0735598922 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 19 |
Hb_011174_020 |
0.0740275639 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 20 |
Hb_002411_110 |
0.0741320065 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC2 [Jatropha curcas] |