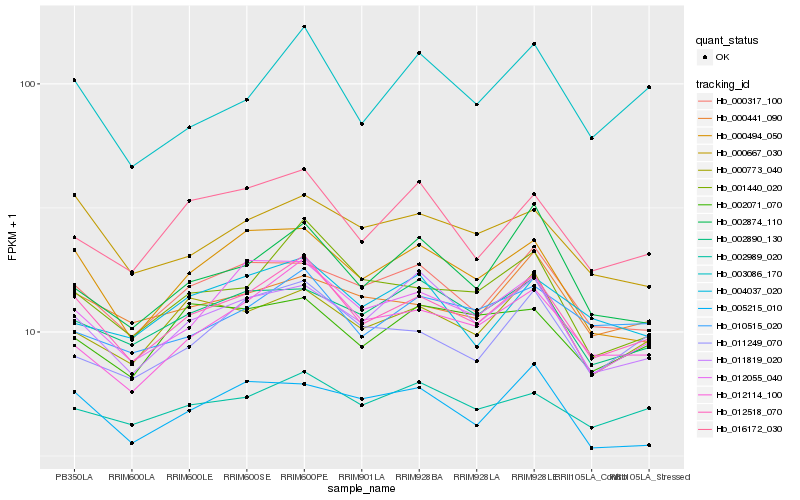
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012518_070 |
0.0 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 9 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001440_020 |
0.0597552391 |
- |
- |
exocyst componenet sec8, putative [Ricinus communis] |
| 3 |
Hb_012055_040 |
0.0603335521 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 4 |
Hb_000317_100 |
0.0606570196 |
- |
- |
PREDICTED: uncharacterized protein LOC105646995 [Jatropha curcas] |
| 5 |
Hb_012114_100 |
0.0617707314 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 6 |
Hb_005215_010 |
0.0641900988 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000494_050 |
0.0646513295 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 8 |
Hb_011819_020 |
0.064683049 |
- |
- |
PREDICTED: uncharacterized protein LOC105643703 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000667_030 |
0.0665322061 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 10 |
Hb_000773_040 |
0.0669001614 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 11 |
Hb_002890_130 |
0.0685734569 |
- |
- |
tip120, putative [Ricinus communis] |
| 12 |
Hb_016172_030 |
0.0703963073 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC [Jatropha curcas] |
| 13 |
Hb_000441_090 |
0.072306412 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 14 |
Hb_002989_020 |
0.0737922699 |
- |
- |
PREDICTED: uncharacterized protein LOC105630013 isoform X2 [Jatropha curcas] |
| 15 |
Hb_011249_070 |
0.07433532 |
- |
- |
hypothetical protein POPTR_0018s12610g [Populus trichocarpa] |
| 16 |
Hb_003086_170 |
0.0745686453 |
- |
- |
PREDICTED: putative phosphatidylglycerol/phosphatidylinositol transfer protein DDB_G0282179 [Jatropha curcas] |
| 17 |
Hb_010515_020 |
0.074849724 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 18 |
Hb_002874_110 |
0.0748666367 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 19 |
Hb_002071_070 |
0.0771528536 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004037_020 |
0.0782888436 |
- |
- |
PREDICTED: uncharacterized protein LOC105632366 isoform X1 [Jatropha curcas] |