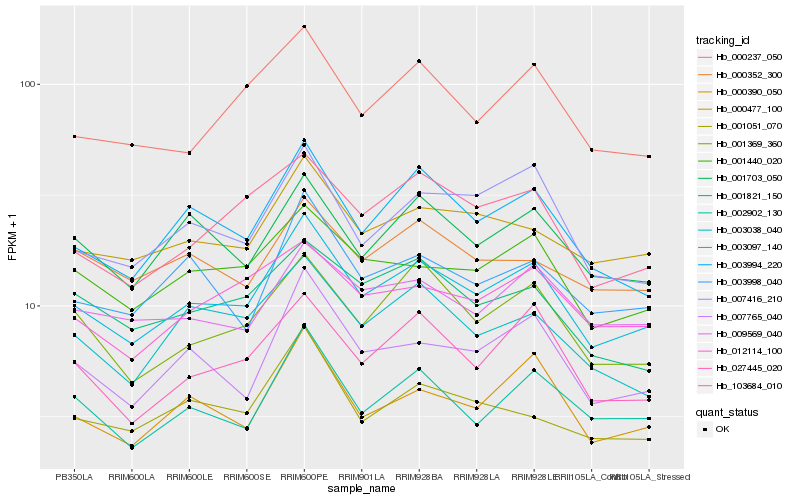
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003097_140 |
0.0 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 2 |
Hb_003994_220 |
0.0677481082 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 3 |
Hb_001440_020 |
0.06822887 |
- |
- |
exocyst componenet sec8, putative [Ricinus communis] |
| 4 |
Hb_007765_040 |
0.0703136707 |
- |
- |
Cytosolic enolase isoform 3 [Theobroma cacao] |
| 5 |
Hb_000477_100 |
0.0712511097 |
- |
- |
PREDICTED: GDP-mannose transporter GONST4 [Jatropha curcas] |
| 6 |
Hb_000390_050 |
0.0723584711 |
- |
- |
PREDICTED: GPCR-type G protein 1 [Jatropha curcas] |
| 7 |
Hb_027445_020 |
0.0731295778 |
- |
- |
PREDICTED: glycosylphosphatidylinositol anchor attachment 1 protein [Jatropha curcas] |
| 8 |
Hb_002902_130 |
0.0738726817 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 9 |
Hb_001369_360 |
0.0740790145 |
- |
- |
embryo yellow protein, partial [Manihot esculenta] |
| 10 |
Hb_001703_050 |
0.0751258684 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 11 |
Hb_007416_210 |
0.0753652218 |
- |
- |
PREDICTED: coatomer subunit beta-1 [Jatropha curcas] |
| 12 |
Hb_000237_050 |
0.0762539411 |
- |
- |
CP2 [Hevea brasiliensis] |
| 13 |
Hb_103684_010 |
0.0772970254 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana tomentosiformis] |
| 14 |
Hb_012114_100 |
0.0790448625 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 15 |
Hb_009569_040 |
0.0791959059 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 16 |
Hb_003998_040 |
0.0804855543 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 17 |
Hb_003038_040 |
0.0829495723 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 18 |
Hb_001821_150 |
0.0836696615 |
- |
- |
PREDICTED: potassium transporter 7-like [Jatropha curcas] |
| 19 |
Hb_001051_070 |
0.0872734013 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000352_300 |
0.0888504193 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3 [Jatropha curcas] |