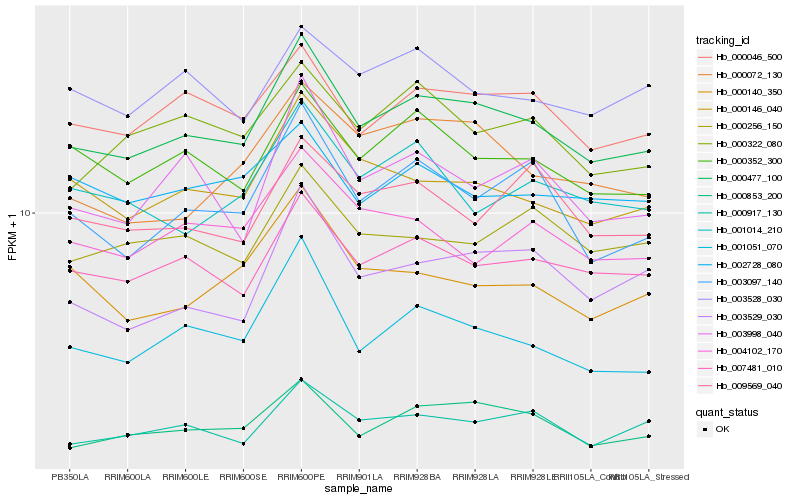
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000477_100 |
0.0 |
- |
- |
PREDICTED: GDP-mannose transporter GONST4 [Jatropha curcas] |
| 2 |
Hb_007481_010 |
0.0577655165 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 3 |
Hb_000046_500 |
0.0639684649 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Jatropha curcas] |
| 4 |
Hb_000352_300 |
0.0653141868 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3 [Jatropha curcas] |
| 5 |
Hb_000146_040 |
0.0659347735 |
- |
- |
PREDICTED: reticulon-like protein B11 [Jatropha curcas] |
| 6 |
Hb_003097_140 |
0.0712511097 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 7 |
Hb_001051_070 |
0.0725351675 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003529_030 |
0.0773591663 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein, putative [Ricinus communis] |
| 9 |
Hb_000256_150 |
0.0797176819 |
- |
- |
PREDICTED: trafficking protein particle complex II-specific subunit 120 homolog [Jatropha curcas] |
| 10 |
Hb_000322_080 |
0.0798466008 |
- |
- |
hypothetical protein JCGZ_07423 [Jatropha curcas] |
| 11 |
Hb_000072_130 |
0.081572344 |
- |
- |
hypothetical protein OsJ_28567 [Oryza sativa Japonica Group] |
| 12 |
Hb_003998_040 |
0.0823918498 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 13 |
Hb_003528_030 |
0.0825381946 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit L-like [Jatropha curcas] |
| 14 |
Hb_004102_170 |
0.0827806387 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 15 |
Hb_001014_210 |
0.0828534699 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 16 |
Hb_000853_200 |
0.082967383 |
- |
- |
PREDICTED: uncharacterized protein LOC105642574 [Jatropha curcas] |
| 17 |
Hb_002728_080 |
0.0836671392 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 18 |
Hb_000140_350 |
0.0837064595 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_009569_040 |
0.0837746266 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 20 |
Hb_000917_130 |
0.0841092763 |
- |
- |
PREDICTED: uncharacterized protein LOC105635730 [Jatropha curcas] |