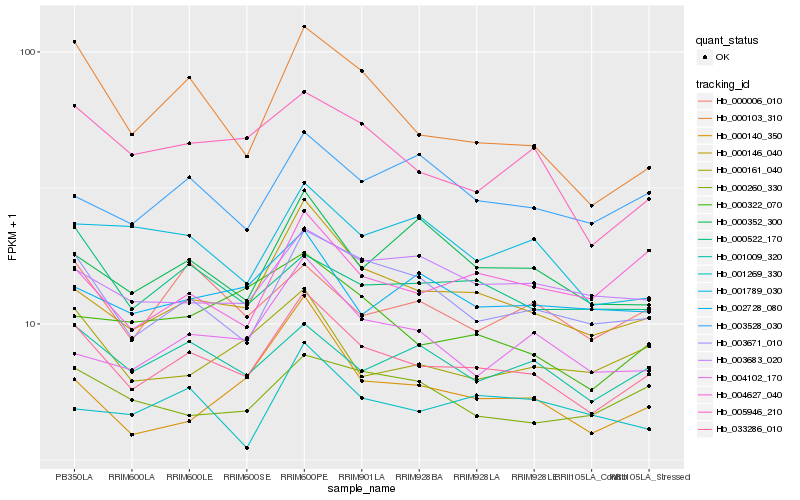
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033286_010 |
0.0 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 14 [Jatropha curcas] |
| 2 |
Hb_000146_040 |
0.0626848714 |
- |
- |
PREDICTED: reticulon-like protein B11 [Jatropha curcas] |
| 3 |
Hb_003671_010 |
0.0650743697 |
- |
- |
PREDICTED: protein transport protein SFT2 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001009_320 |
0.0734897433 |
- |
- |
PREDICTED: uncharacterized protein LOC105640250 [Jatropha curcas] |
| 5 |
Hb_005946_210 |
0.0784349477 |
- |
- |
hypothetical protein JCGZ_07228 [Jatropha curcas] |
| 6 |
Hb_000522_170 |
0.0810820255 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 1 [Jatropha curcas] |
| 7 |
Hb_000006_010 |
0.0812301637 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003683_020 |
0.0829709162 |
- |
- |
serine/threonine protein kinase, putative [Ricinus communis] |
| 9 |
Hb_003528_030 |
0.0852820863 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit L-like [Jatropha curcas] |
| 10 |
Hb_000260_330 |
0.0859853665 |
- |
- |
PREDICTED: uncharacterized protein LOC103438625 [Malus domestica] |
| 11 |
Hb_000103_310 |
0.0860988944 |
- |
- |
PREDICTED: transmembrane emp24 domain-containing protein p24beta3 [Jatropha curcas] |
| 12 |
Hb_002728_080 |
0.086616923 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 13 |
Hb_000352_300 |
0.0866908629 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3 [Jatropha curcas] |
| 14 |
Hb_000161_040 |
0.0872718093 |
- |
- |
PREDICTED: protein EI24 homolog [Jatropha curcas] |
| 15 |
Hb_000140_350 |
0.087562559 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004102_170 |
0.0883778792 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 17 |
Hb_004627_040 |
0.0887592179 |
- |
- |
hypothetical protein POPTR_0010s22770g [Populus trichocarpa] |
| 18 |
Hb_000322_070 |
0.088802996 |
- |
- |
PREDICTED: glycylpeptide N-tetradecanoyltransferase 1 [Jatropha curcas] |
| 19 |
Hb_001269_330 |
0.0896108031 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 20 |
Hb_001789_030 |
0.0896447111 |
- |
- |
PREDICTED: signal peptide peptidase-like 1 [Jatropha curcas] |