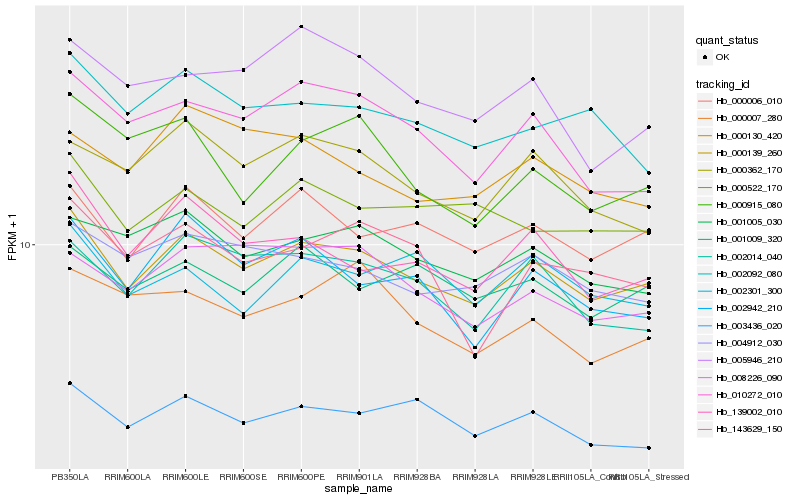
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000139_260 |
0.0 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 2 |
Hb_000006_010 |
0.0604753643 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002014_040 |
0.0725021578 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 4 |
Hb_001005_030 |
0.0729749367 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 5 |
Hb_004912_030 |
0.0732050852 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 6 |
Hb_139002_010 |
0.0738006851 |
- |
- |
alpha-(1,4)-fucosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_010272_010 |
0.0752293977 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 8 |
Hb_000522_170 |
0.0759881226 |
- |
- |
PREDICTED: phosphatidylinositol:ceramide inositolphosphotransferase 1 [Jatropha curcas] |
| 9 |
Hb_001009_320 |
0.0765079735 |
- |
- |
PREDICTED: uncharacterized protein LOC105640250 [Jatropha curcas] |
| 10 |
Hb_002092_080 |
0.0771658086 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A beta isoform-like [Jatropha curcas] |
| 11 |
Hb_003436_020 |
0.0777269243 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105636276 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000362_170 |
0.0779475189 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 13 |
Hb_002942_210 |
0.0793169412 |
- |
- |
alpha-2,8-sialyltransferase 8b, putative [Ricinus communis] |
| 14 |
Hb_008226_090 |
0.0830562126 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002301_300 |
0.0853759363 |
- |
- |
PREDICTED: dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase isoform X1 [Jatropha curcas] |
| 16 |
Hb_143629_150 |
0.0864457177 |
- |
- |
PREDICTED: flap endonuclease 1 isoform X2 [Prunus mume] |
| 17 |
Hb_005946_210 |
0.0870935719 |
- |
- |
hypothetical protein JCGZ_07228 [Jatropha curcas] |
| 18 |
Hb_000915_080 |
0.0874503754 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 19 |
Hb_000007_280 |
0.0883359672 |
- |
- |
hypothetical protein VITISV_000066 [Vitis vinifera] |
| 20 |
Hb_000130_420 |
0.089289718 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |