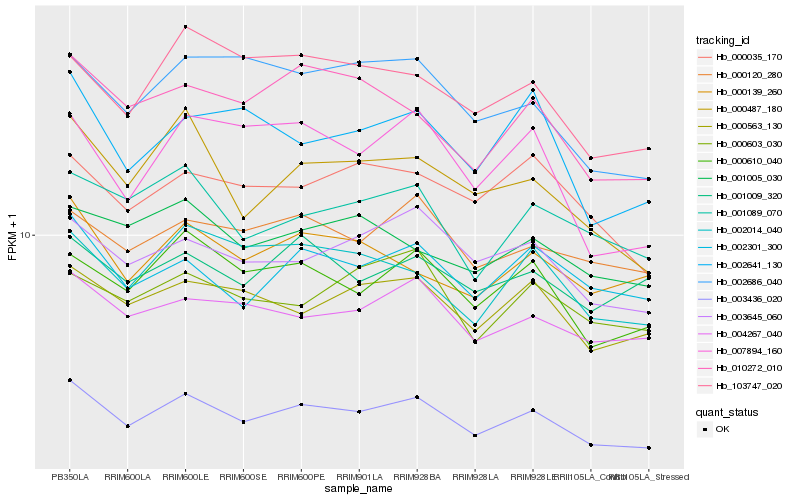
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003436_020 |
0.0 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105636276 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000610_040 |
0.0639004696 |
- |
- |
PREDICTED: uncharacterized protein LOC105635307 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003645_060 |
0.0664883256 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_004267_040 |
0.0691265048 |
- |
- |
PREDICTED: transcriptional adapter ADA2a [Jatropha curcas] |
| 5 |
Hb_000487_180 |
0.0691982488 |
- |
- |
PREDICTED: post-GPI attachment to proteins factor 3 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000563_130 |
0.0692138331 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 7 |
Hb_002301_300 |
0.0700489073 |
- |
- |
PREDICTED: dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase isoform X1 [Jatropha curcas] |
| 8 |
Hb_000120_280 |
0.073434643 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 9 |
Hb_002014_040 |
0.0736106012 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 10 |
Hb_010272_010 |
0.07434909 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 11 |
Hb_001089_070 |
0.075624968 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 3 [Jatropha curcas] |
| 12 |
Hb_002686_040 |
0.0757304023 |
- |
- |
PREDICTED: T-complex protein 1 subunit theta [Jatropha curcas] |
| 13 |
Hb_103747_020 |
0.076499474 |
- |
- |
calcium dependent protein kinase, partial [Hevea brasiliensis] |
| 14 |
Hb_000139_260 |
0.0777269243 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 15 |
Hb_000035_170 |
0.0785579664 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 2-like [Jatropha curcas] |
| 16 |
Hb_007894_160 |
0.0792246811 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 17 |
Hb_001005_030 |
0.0808521776 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 18 |
Hb_001009_320 |
0.0820623459 |
- |
- |
PREDICTED: uncharacterized protein LOC105640250 [Jatropha curcas] |
| 19 |
Hb_002641_130 |
0.0828866493 |
- |
- |
PREDICTED: uncharacterized protein LOC105643556 [Jatropha curcas] |
| 20 |
Hb_000603_030 |
0.0833852738 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |