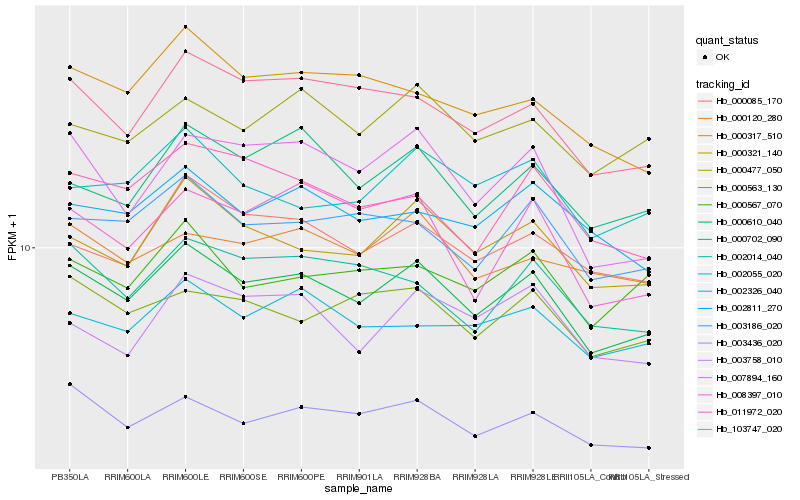
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000610_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635307 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000321_140 |
0.0633556155 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 3 |
Hb_003436_020 |
0.0639004696 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105636276 isoform X2 [Jatropha curcas] |
| 4 |
Hb_103747_020 |
0.0640430087 |
- |
- |
calcium dependent protein kinase, partial [Hevea brasiliensis] |
| 5 |
Hb_003186_020 |
0.0680539543 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007894_160 |
0.0687656555 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 7 |
Hb_000702_090 |
0.0714585071 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 8 |
Hb_000085_170 |
0.072577097 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000563_130 |
0.0759705082 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 10 |
Hb_002014_040 |
0.0762310882 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 11 |
Hb_002326_040 |
0.0763105223 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 12 |
Hb_011972_020 |
0.0763607688 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 13 |
Hb_000477_050 |
0.0781517816 |
- |
- |
PREDICTED: 26S protease regulatory subunit 8 homolog A [Jatropha curcas] |
| 14 |
Hb_000317_510 |
0.0785174084 |
- |
- |
PREDICTED: long chain base biosynthesis protein 1 [Jatropha curcas] |
| 15 |
Hb_002811_270 |
0.0787932697 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 16 |
Hb_000120_280 |
0.0800216355 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 17 |
Hb_002055_020 |
0.0800463262 |
- |
- |
PREDICTED: large subunit GTPase 1 homolog [Jatropha curcas] |
| 18 |
Hb_000567_070 |
0.0805278149 |
- |
- |
autophagy protein, putative [Ricinus communis] |
| 19 |
Hb_008397_010 |
0.0807285284 |
- |
- |
PREDICTED: uncharacterized protein LOC105640192 isoform X2 [Jatropha curcas] |
| 20 |
Hb_003758_010 |
0.0824742132 |
transcription factor |
TF Family: VOZ |
PREDICTED: transcription factor VOZ1 isoform X1 [Jatropha curcas] |