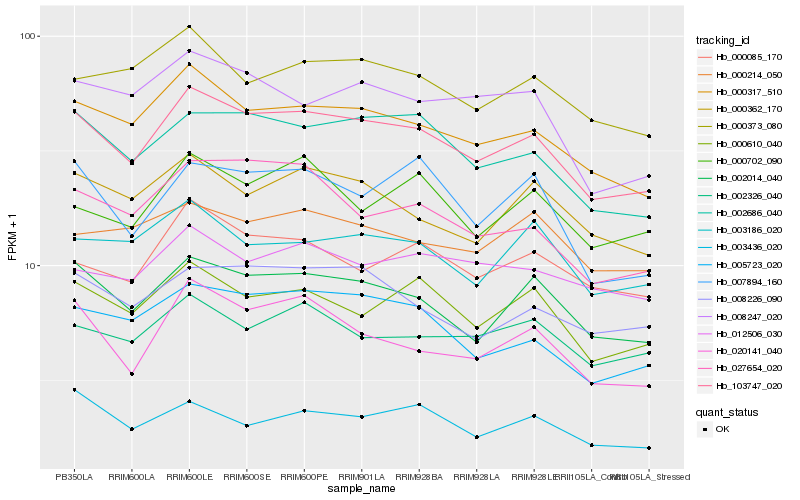
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_103747_020 |
0.0 |
- |
- |
calcium dependent protein kinase, partial [Hevea brasiliensis] |
| 2 |
Hb_000317_510 |
0.0493504376 |
- |
- |
PREDICTED: long chain base biosynthesis protein 1 [Jatropha curcas] |
| 3 |
Hb_002014_040 |
0.0516399335 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 4 |
Hb_002686_040 |
0.0557019717 |
- |
- |
PREDICTED: T-complex protein 1 subunit theta [Jatropha curcas] |
| 5 |
Hb_005723_020 |
0.0629308177 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000610_040 |
0.0640430087 |
- |
- |
PREDICTED: uncharacterized protein LOC105635307 isoform X1 [Jatropha curcas] |
| 7 |
Hb_008226_090 |
0.0679361118 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 8 |
Hb_020141_040 |
0.0694008216 |
transcription factor |
TF Family: STAT |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000362_170 |
0.0703344209 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 10 |
Hb_000085_170 |
0.0718978021 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_002326_040 |
0.0737402132 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 12 |
Hb_000373_080 |
0.0738928227 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 13 |
Hb_027654_020 |
0.074164777 |
- |
- |
PREDICTED: LIMR family protein At5g01460 [Jatropha curcas] |
| 14 |
Hb_003186_020 |
0.0759893453 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 15 |
Hb_003436_020 |
0.076499474 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105636276 isoform X2 [Jatropha curcas] |
| 16 |
Hb_008247_020 |
0.0768542411 |
- |
- |
PREDICTED: cyclin-L1-1-like [Jatropha curcas] |
| 17 |
Hb_007894_160 |
0.0773006139 |
- |
- |
PREDICTED: purple acid phosphatase 18 [Jatropha curcas] |
| 18 |
Hb_000702_090 |
0.0779120389 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 19 |
Hb_000214_050 |
0.079002806 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_012506_030 |
0.0791247279 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |