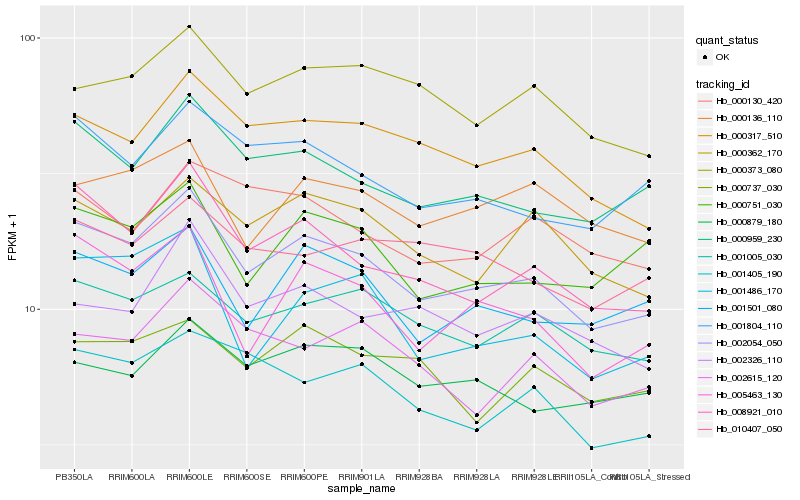
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002054_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 2 |
Hb_008921_010 |
0.0566998519 |
- |
- |
PREDICTED: uncharacterized protein LOC105629323 [Jatropha curcas] |
| 3 |
Hb_000959_230 |
0.0637621822 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 12 homolog A [Populus euphratica] |
| 4 |
Hb_000317_510 |
0.0672801871 |
- |
- |
PREDICTED: long chain base biosynthesis protein 1 [Jatropha curcas] |
| 5 |
Hb_005463_130 |
0.0704543775 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 6 |
Hb_001005_030 |
0.0715960385 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 7 |
Hb_001501_080 |
0.0716497801 |
- |
- |
PREDICTED: uncharacterized protein LOC105640832 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001804_110 |
0.0729383361 |
- |
- |
hypothetical protein CISIN_1g031200mg [Citrus sinensis] |
| 9 |
Hb_000362_170 |
0.0760509002 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 10 |
Hb_001405_190 |
0.0780330532 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000751_030 |
0.0794251791 |
- |
- |
Vesicle transport protein GOT1B, putative [Ricinus communis] |
| 12 |
Hb_000373_080 |
0.0809785102 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 13 |
Hb_002326_110 |
0.0834429019 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |
| 14 |
Hb_001486_170 |
0.0839094888 |
- |
- |
PREDICTED: syntaxin-132 [Jatropha curcas] |
| 15 |
Hb_000136_110 |
0.0841373008 |
- |
- |
hypothetical protein B456_011G031000, partial [Gossypium raimondii] |
| 16 |
Hb_000879_180 |
0.0841626186 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 17 |
Hb_002615_120 |
0.0844108769 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000130_420 |
0.0845312722 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 19 |
Hb_010407_050 |
0.0847259734 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 20 |
Hb_000737_030 |
0.0854195489 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |