| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
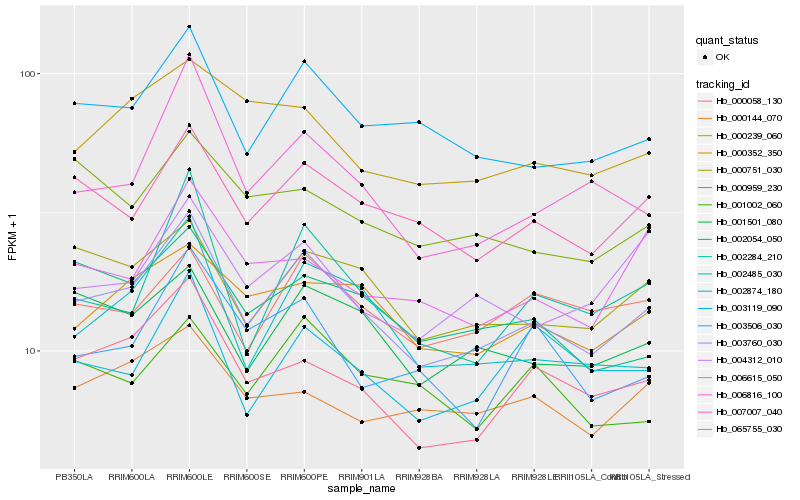
Hb_003760_030 |
0.0 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 2 |
Hb_003119_090 |
0.0709197042 |
- |
- |
PREDICTED: 14-3-3 protein 6 [Jatropha curcas] |
| 3 |
Hb_007007_040 |
0.0714037873 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 4 |
Hb_065755_030 |
0.0775451844 |
- |
- |
- |
| 5 |
Hb_000751_030 |
0.0776983952 |
- |
- |
Vesicle transport protein GOT1B, putative [Ricinus communis] |
| 6 |
Hb_001501_080 |
0.0801604866 |
- |
- |
PREDICTED: uncharacterized protein LOC105640832 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002485_030 |
0.081815255 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase 1-like [Jatropha curcas] |
| 8 |
Hb_000239_060 |
0.0836559448 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_006816_100 |
0.0858303496 |
- |
- |
hypothetical protein CISIN_1g0373181mg, partial [Citrus sinensis] |
| 10 |
Hb_002284_210 |
0.0870094822 |
- |
- |
PREDICTED: protein canopy-1 [Jatropha curcas] |
| 11 |
Hb_002054_050 |
0.0895846911 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 12 |
Hb_000058_130 |
0.0905604204 |
- |
- |
PREDICTED: uncharacterized protein LOC105640884 [Jatropha curcas] |
| 13 |
Hb_000352_350 |
0.0906715996 |
- |
- |
PREDICTED: monothiol glutaredoxin-S7, chloroplastic [Jatropha curcas] |
| 14 |
Hb_004312_010 |
0.0926189017 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X2 [Jatropha curcas] |
| 15 |
Hb_002874_180 |
0.0943922596 |
- |
- |
PREDICTED: protein AIG2-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_006615_050 |
0.095802022 |
- |
- |
PREDICTED: uncharacterized protein LOC105640686 [Jatropha curcas] |
| 17 |
Hb_000959_230 |
0.0962603518 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 12 homolog A [Populus euphratica] |
| 18 |
Hb_000144_070 |
0.0976233288 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003506_030 |
0.0976710409 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial isoform X2 [Jatropha curcas] |
| 20 |
Hb_001002_060 |
0.0980193654 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |