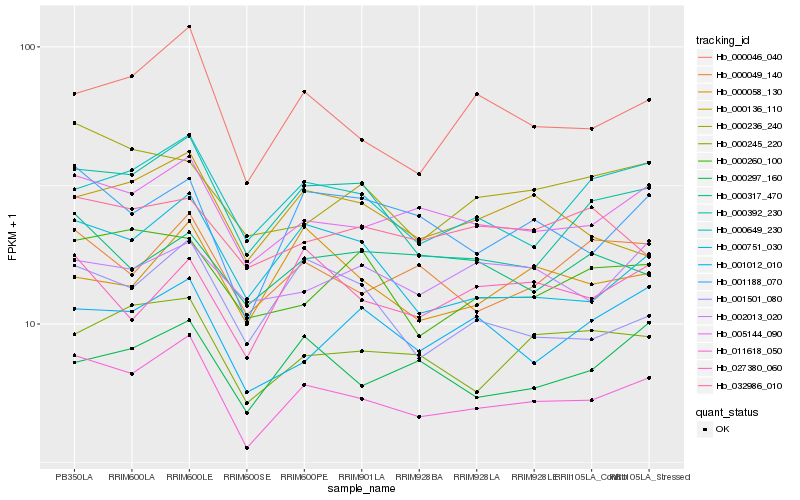
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011618_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634948 [Jatropha curcas] |
| 2 |
Hb_005144_090 |
0.0475334975 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 24 homolog 1-like [Citrus sinensis] |
| 3 |
Hb_000046_040 |
0.0699983152 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000649_230 |
0.0731445227 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 variant 1A-like [Jatropha curcas] |
| 5 |
Hb_000751_030 |
0.0734677417 |
- |
- |
Vesicle transport protein GOT1B, putative [Ricinus communis] |
| 6 |
Hb_000049_140 |
0.0741965931 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 9, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000245_220 |
0.074676011 |
- |
- |
PREDICTED: uncharacterized protein LOC105635566 [Jatropha curcas] |
| 8 |
Hb_000392_230 |
0.0756587987 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001188_070 |
0.0770189281 |
- |
- |
PREDICTED: protein disulfide-isomerase 5-1 [Jatropha curcas] |
| 10 |
Hb_001501_080 |
0.0820961987 |
- |
- |
PREDICTED: uncharacterized protein LOC105640832 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000297_160 |
0.0827965587 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |
| 12 |
Hb_000260_100 |
0.0853660146 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000136_110 |
0.0854351692 |
- |
- |
hypothetical protein B456_011G031000, partial [Gossypium raimondii] |
| 14 |
Hb_001012_010 |
0.086545957 |
- |
- |
PREDICTED: XIAP-associated factor 1 [Jatropha curcas] |
| 15 |
Hb_000236_240 |
0.0873990726 |
- |
- |
Pre-mRNA-splicing factor cwc15, putative [Ricinus communis] |
| 16 |
Hb_000317_470 |
0.0878907969 |
- |
- |
PREDICTED: CCR4-NOT transcription complex subunit 11 [Jatropha curcas] |
| 17 |
Hb_032986_010 |
0.0882241199 |
- |
- |
Nicotinate-nucleotide pyrophosphorylase [carboxylating], putative [Ricinus communis] |
| 18 |
Hb_002013_020 |
0.0883352911 |
- |
- |
PREDICTED: uncharacterized protein LOC105641613 [Jatropha curcas] |
| 19 |
Hb_027380_060 |
0.0884895145 |
- |
- |
PREDICTED: putative ALA-interacting subunit 2 [Jatropha curcas] |
| 20 |
Hb_000058_130 |
0.0885071417 |
- |
- |
PREDICTED: uncharacterized protein LOC105640884 [Jatropha curcas] |