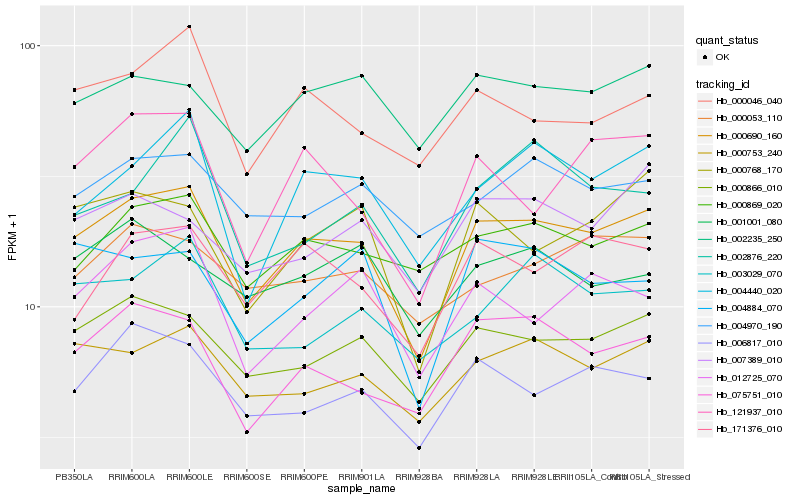
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000690_160 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105121500 [Populus euphratica] |
| 2 |
Hb_000866_010 |
0.0738658799 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000753_240 |
0.0825501302 |
- |
- |
PREDICTED: ADP,ATP carrier protein ER-ANT1 [Jatropha curcas] |
| 4 |
Hb_075751_010 |
0.0839536198 |
- |
- |
PREDICTED: uncharacterized protein LOC105635783 isoform X2 [Jatropha curcas] |
| 5 |
Hb_006817_010 |
0.0885612335 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004440_020 |
0.0889327963 |
- |
- |
AP-3 complex subunit sigma-1, putative [Ricinus communis] |
| 7 |
Hb_000768_170 |
0.0891929997 |
- |
- |
hypothetical protein JCGZ_02624 [Jatropha curcas] |
| 8 |
Hb_171376_010 |
0.0905492804 |
- |
- |
PREDICTED: uncharacterized protein LOC105630877 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000046_040 |
0.0923386045 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000869_020 |
0.0924412472 |
- |
- |
PREDICTED: uncharacterized protein LOC105641036 [Jatropha curcas] |
| 11 |
Hb_002235_250 |
0.0929639542 |
- |
- |
PREDICTED: cystathionine gamma-synthase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_121937_010 |
0.093328536 |
- |
- |
hypothetical protein B456_013G261900 [Gossypium raimondii] |
| 13 |
Hb_004884_070 |
0.0944234452 |
- |
- |
PREDICTED: ethylene-overproduction protein 1 [Jatropha curcas] |
| 14 |
Hb_012725_070 |
0.096074931 |
- |
- |
PREDICTED: uncharacterized protein LOC105636534 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000053_110 |
0.0974318366 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004970_190 |
0.0974598137 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_007389_010 |
0.099269398 |
- |
- |
PREDICTED: THO complex subunit 7A [Jatropha curcas] |
| 18 |
Hb_003029_070 |
0.1018405728 |
- |
- |
PREDICTED: beta-carotene isomerase D27, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_001001_080 |
0.1033081628 |
- |
- |
PREDICTED: uncharacterized protein LOC105633661 [Jatropha curcas] |
| 20 |
Hb_002876_220 |
0.1051522812 |
- |
- |
PREDICTED: uncharacterized protein LOC105633932 [Jatropha curcas] |