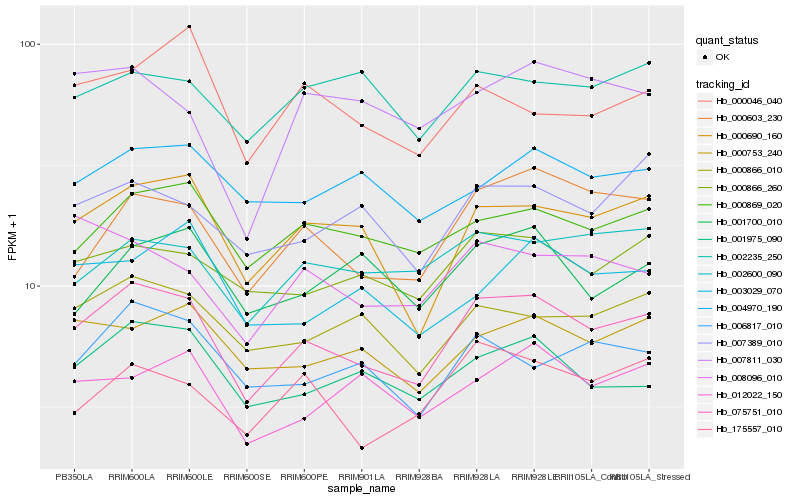
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_075751_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105635783 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000690_160 |
0.0839536198 |
- |
- |
PREDICTED: uncharacterized protein LOC105121500 [Populus euphratica] |
| 3 |
Hb_000603_230 |
0.0924403493 |
- |
- |
copper-transporting atpase paa1, putative [Ricinus communis] |
| 4 |
Hb_001975_090 |
0.0927934048 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 5 |
Hb_000869_020 |
0.0957817625 |
- |
- |
PREDICTED: uncharacterized protein LOC105641036 [Jatropha curcas] |
| 6 |
Hb_000866_010 |
0.0960838005 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000866_260 |
0.0976424228 |
- |
- |
PREDICTED: F-box/kelch-repeat protein SKIP4 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000753_240 |
0.1014110487 |
- |
- |
PREDICTED: ADP,ATP carrier protein ER-ANT1 [Jatropha curcas] |
| 9 |
Hb_006817_010 |
0.1024177517 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002600_090 |
0.1044033679 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_175557_010 |
0.1044044302 |
- |
- |
glutamate-1-semialdehyde 2,1-aminomutase, putative [Ricinus communis] |
| 12 |
Hb_007389_010 |
0.1068504636 |
- |
- |
PREDICTED: THO complex subunit 7A [Jatropha curcas] |
| 13 |
Hb_012022_150 |
0.1094359549 |
- |
- |
PREDICTED: protein root UVB sensitive 5 [Jatropha curcas] |
| 14 |
Hb_004970_190 |
0.1119360676 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_008096_010 |
0.112441596 |
- |
- |
hypothetical protein MTR_1g045660 [Medicago truncatula] |
| 16 |
Hb_002235_250 |
0.1137190196 |
- |
- |
PREDICTED: cystathionine gamma-synthase 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_003029_070 |
0.1143011813 |
- |
- |
PREDICTED: beta-carotene isomerase D27, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_000046_040 |
0.1144764825 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_007811_030 |
0.1152825997 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_001700_010 |
0.1158022589 |
- |
- |
PREDICTED: GTP-binding protein At2g22870 [Jatropha curcas] |