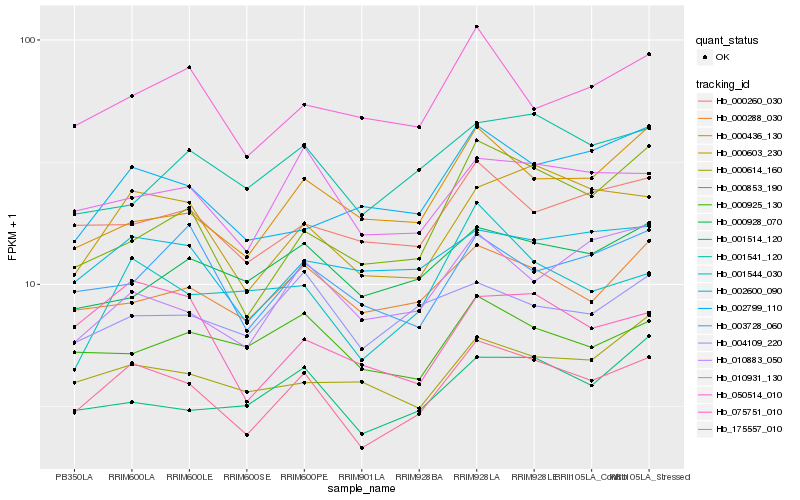
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_175557_010 |
0.0 |
- |
- |
glutamate-1-semialdehyde 2,1-aminomutase, putative [Ricinus communis] |
| 2 |
Hb_000603_230 |
0.0814442089 |
- |
- |
copper-transporting atpase paa1, putative [Ricinus communis] |
| 3 |
Hb_010931_130 |
0.0895690848 |
- |
- |
PREDICTED: mitogen-activated protein kinase 20 [Jatropha curcas] |
| 4 |
Hb_002600_090 |
0.1005239767 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_000288_030 |
0.1023706045 |
- |
- |
PREDICTED: syntaxin-32 [Jatropha curcas] |
| 6 |
Hb_000260_030 |
0.1024602992 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 7 |
Hb_001514_120 |
0.1025641385 |
- |
- |
hypothetical protein VITISV_015004 [Vitis vinifera] |
| 8 |
Hb_075751_010 |
0.1044044302 |
- |
- |
PREDICTED: uncharacterized protein LOC105635783 isoform X2 [Jatropha curcas] |
| 9 |
Hb_050514_010 |
0.1097275964 |
- |
- |
hypothetical protein 29 [Hevea brasiliensis] |
| 10 |
Hb_001544_030 |
0.1103499707 |
- |
- |
PREDICTED: putative 1-phosphatidylinositol-3-phosphate 5-kinase FAB1C [Jatropha curcas] |
| 11 |
Hb_004109_220 |
0.1113823352 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 9-like [Populus euphratica] |
| 12 |
Hb_000925_130 |
0.1123183683 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At3g05675-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_001541_120 |
0.1141130767 |
- |
- |
hypothetical protein JCGZ_17842 [Jatropha curcas] |
| 14 |
Hb_002799_110 |
0.1143065994 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 15 |
Hb_000853_190 |
0.1146641771 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 16 |
Hb_010883_050 |
0.1152171972 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 1C-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000436_130 |
0.1154943389 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |
| 18 |
Hb_000928_070 |
0.1155439901 |
- |
- |
PREDICTED: maltose excess protein 1, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_003728_060 |
0.1155726195 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 4-like [Jatropha curcas] |
| 20 |
Hb_000614_160 |
0.1155982578 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04130, mitochondrial [Jatropha curcas] |