| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
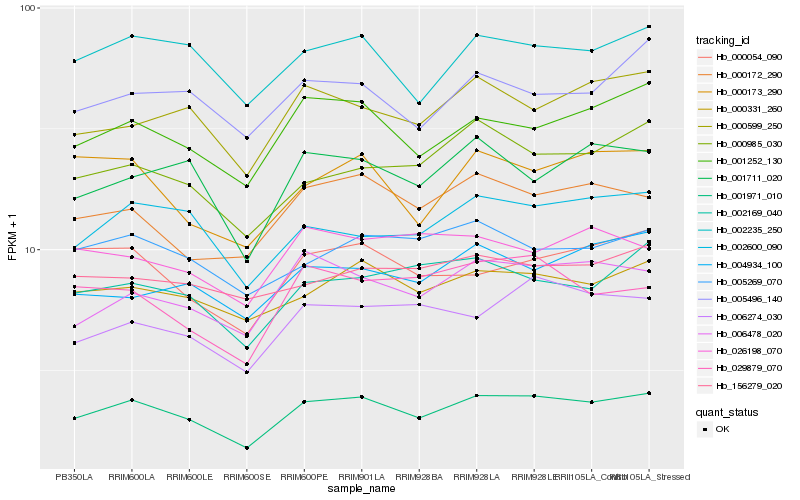
Hb_001971_010 |
0.0 |
- |
- |
F23N19.4 [Arabidopsis thaliana] |
| 2 |
Hb_001252_130 |
0.0588422756 |
- |
- |
PREDICTED: protein YIPF1 homolog [Jatropha curcas] |
| 3 |
Hb_000331_260 |
0.0623333495 |
- |
- |
PREDICTED: WD repeat-containing protein 74 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002235_250 |
0.064223752 |
- |
- |
PREDICTED: cystathionine gamma-synthase 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002600_090 |
0.0681632846 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 6 |
Hb_006478_020 |
0.0686444557 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |
| 7 |
Hb_000172_290 |
0.0687198628 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 12 [Jatropha curcas] |
| 8 |
Hb_000054_090 |
0.07113648 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 9 |
Hb_001711_020 |
0.0715888803 |
- |
- |
PREDICTED: AP-2 complex subunit mu [Jatropha curcas] |
| 10 |
Hb_005269_070 |
0.0719983371 |
- |
- |
PREDICTED: translation factor GUF1 homolog, organellar chromatophore [Jatropha curcas] |
| 11 |
Hb_000599_250 |
0.0720210859 |
- |
- |
hypothetical protein POPTR_0002s03730g [Populus trichocarpa] |
| 12 |
Hb_002169_040 |
0.0721816784 |
- |
- |
unknown [Populus trichocarpa] |
| 13 |
Hb_000985_030 |
0.0724002505 |
- |
- |
PREDICTED: RRP15-like protein [Populus euphratica] |
| 14 |
Hb_006274_030 |
0.0738797438 |
- |
- |
F-box and wd40 domain protein, putative [Ricinus communis] |
| 15 |
Hb_005496_140 |
0.0767949266 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000173_290 |
0.0782129687 |
- |
- |
guanylate kinase, putative [Ricinus communis] |
| 17 |
Hb_029879_070 |
0.0782347267 |
- |
- |
PREDICTED: phospholipase A I [Jatropha curcas] |
| 18 |
Hb_156279_020 |
0.0796582573 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 19 |
Hb_004934_100 |
0.0800209796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_026198_070 |
0.0815112046 |
- |
- |
PREDICTED: uncharacterized membrane protein At4g09580 [Jatropha curcas] |