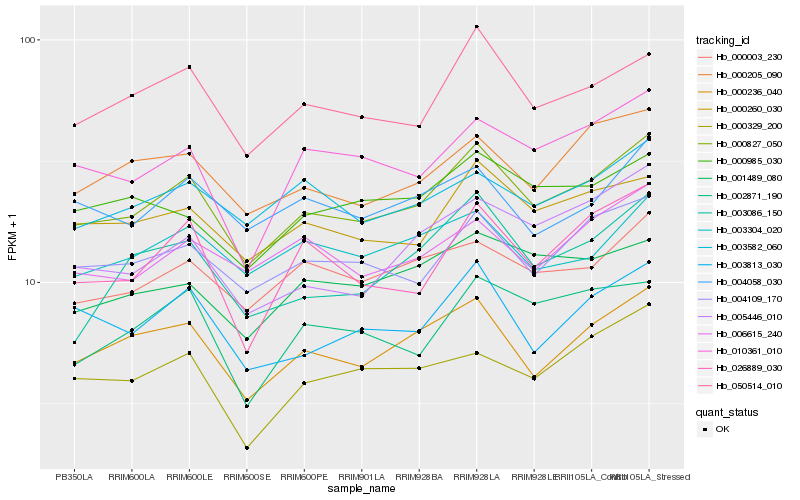
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000827_050 |
0.0 |
- |
- |
PREDICTED: tyrosine--tRNA ligase, cytoplasmic-like [Jatropha curcas] |
| 2 |
Hb_000236_040 |
0.0566251233 |
- |
- |
PREDICTED: uncharacterized protein LOC105633150 isoform X1 [Jatropha curcas] |
| 3 |
Hb_050514_010 |
0.0581797078 |
- |
- |
hypothetical protein 29 [Hevea brasiliensis] |
| 4 |
Hb_004109_170 |
0.0615838826 |
- |
- |
PREDICTED: probable ribokinase [Jatropha curcas] |
| 5 |
Hb_000260_030 |
0.0634075482 |
transcription factor |
TF Family: IWS1 |
transcription elongation factor s-II, putative [Ricinus communis] |
| 6 |
Hb_000003_230 |
0.0667910975 |
- |
- |
PREDICTED: FAD synthetase 2, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000205_090 |
0.0683638508 |
- |
- |
hypothetical protein POPTR_0019s03570g [Populus trichocarpa] |
| 8 |
Hb_003813_030 |
0.0699987596 |
- |
- |
PREDICTED: mannose-6-phosphate isomerase 1 [Jatropha curcas] |
| 9 |
Hb_010361_010 |
0.0716294688 |
- |
- |
PREDICTED: ras-related protein RABH1b [Jatropha curcas] |
| 10 |
Hb_026889_030 |
0.0734027269 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_006615_240 |
0.0745486402 |
- |
- |
PREDICTED: uncharacterized protein LOC105640673 [Jatropha curcas] |
| 12 |
Hb_003582_060 |
0.0746283135 |
transcription factor |
TF Family: BBR-BPC |
PREDICTED: protein BASIC PENTACYSTEINE2-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_003304_020 |
0.0762854751 |
- |
- |
Polyadenylate-binding protein RBP47C [Glycine soja] |
| 14 |
Hb_001489_080 |
0.0768231345 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_004058_030 |
0.0771021493 |
- |
- |
PREDICTED: stomatin-like protein 2, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000985_030 |
0.0776485104 |
- |
- |
PREDICTED: RRP15-like protein [Populus euphratica] |
| 17 |
Hb_003086_150 |
0.0787964534 |
- |
- |
PREDICTED: uncharacterized protein LOC105635789 [Jatropha curcas] |
| 18 |
Hb_005446_010 |
0.0795366416 |
- |
- |
PREDICTED: zinc finger protein 830 [Jatropha curcas] |
| 19 |
Hb_000329_200 |
0.0799659238 |
- |
- |
PREDICTED: uncharacterized protein LOC105639316 [Jatropha curcas] |
| 20 |
Hb_002871_190 |
0.0801456278 |
- |
- |
PREDICTED: uncharacterized protein LOC105628867 [Jatropha curcas] |