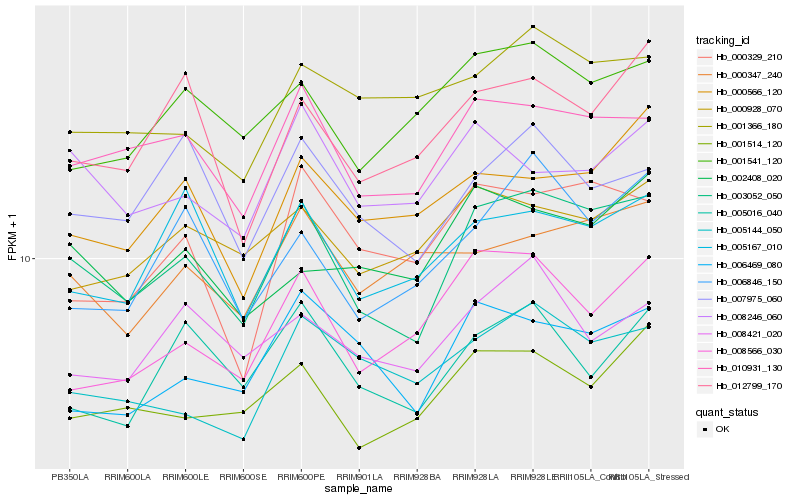
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003052_050 |
0.0 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 2 |
Hb_010931_130 |
0.0796690499 |
- |
- |
PREDICTED: mitogen-activated protein kinase 20 [Jatropha curcas] |
| 3 |
Hb_001514_120 |
0.0971467228 |
- |
- |
hypothetical protein VITISV_015004 [Vitis vinifera] |
| 4 |
Hb_005167_010 |
0.1050734033 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X1 [Jatropha curcas] |
| 5 |
Hb_012799_170 |
0.1065053904 |
- |
- |
PREDICTED: protein GLC8 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001541_120 |
0.106878835 |
- |
- |
hypothetical protein JCGZ_17842 [Jatropha curcas] |
| 7 |
Hb_007975_060 |
0.1069794435 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |
| 8 |
Hb_006469_080 |
0.1070385019 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_008246_060 |
0.1088466561 |
- |
- |
hypothetical protein F383_01577 [Gossypium arboreum] |
| 10 |
Hb_005144_050 |
0.1088583885 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 isoform X2 [Vitis vinifera] |
| 11 |
Hb_008566_030 |
0.1095625696 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 12 |
Hb_000928_070 |
0.1096993541 |
- |
- |
PREDICTED: maltose excess protein 1, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_000329_210 |
0.1103752924 |
- |
- |
Golgi snare 12 isoform 1 [Theobroma cacao] |
| 14 |
Hb_006846_150 |
0.1107274616 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000566_120 |
0.1109310565 |
- |
- |
PREDICTED: uncharacterized protein LOC105123035 isoform X1 [Populus euphratica] |
| 16 |
Hb_002408_020 |
0.111631717 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X4 [Jatropha curcas] |
| 17 |
Hb_000347_240 |
0.1120333714 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Pyrus x bretschneideri] |
| 18 |
Hb_005016_040 |
0.1128454994 |
transcription factor |
TF Family: E2F-DP |
PREDICTED: transcription factor-like protein DPB isoform X2 [Jatropha curcas] |
| 19 |
Hb_001366_180 |
0.1138549893 |
- |
- |
PREDICTED: translocase of chloroplast 33, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_008421_020 |
0.1140608529 |
- |
- |
PREDICTED: uncharacterized protein LOC105635546 isoform X2 [Jatropha curcas] |