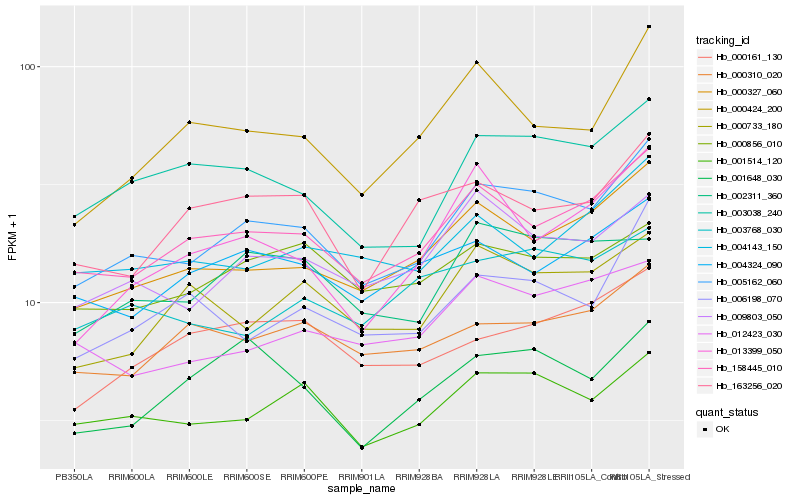
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005162_060 |
0.0 |
- |
- |
PREDICTED: putative septum site-determining protein minD homolog, chloroplastic [Jatropha curcas] |
| 2 |
Hb_158445_010 |
0.0679436611 |
- |
- |
PREDICTED: cysteine protease ATG4-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_000327_060 |
0.0742983016 |
- |
- |
PREDICTED: uncharacterized protein LOC105635258 isoform X2 [Jatropha curcas] |
| 4 |
Hb_009803_050 |
0.0772911744 |
- |
- |
PREDICTED: uncharacterized protein LOC105644460 [Jatropha curcas] |
| 5 |
Hb_001514_120 |
0.0797058957 |
- |
- |
hypothetical protein VITISV_015004 [Vitis vinifera] |
| 6 |
Hb_003038_240 |
0.0828986042 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000424_200 |
0.0968248132 |
- |
- |
hypothetical protein JCGZ_07583 [Jatropha curcas] |
| 8 |
Hb_006198_070 |
0.0996630203 |
- |
- |
pantoate-beta-alanine ligase, putative [Ricinus communis] |
| 9 |
Hb_012423_030 |
0.1002929918 |
- |
- |
PREDICTED: probable sodium/metabolite cotransporter BASS6, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_002311_360 |
0.1004362507 |
- |
- |
PREDICTED: uncharacterized protein LOC105114273 [Populus euphratica] |
| 11 |
Hb_000310_020 |
0.1020275839 |
- |
- |
hypothetical protein JCGZ_20797 [Jatropha curcas] |
| 12 |
Hb_001648_030 |
0.1026720321 |
- |
- |
PREDICTED: uncharacterized protein LOC105628931 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000733_180 |
0.1030213829 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein B-like [Jatropha curcas] |
| 14 |
Hb_004143_150 |
0.1034250938 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 24-like isoform X6 [Jatropha curcas] |
| 15 |
Hb_000856_010 |
0.1038301092 |
- |
- |
PREDICTED: uncharacterized protein LOC105640466 [Jatropha curcas] |
| 16 |
Hb_004324_090 |
0.104290031 |
- |
- |
PREDICTED: uncharacterized protein LOC105648352 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000161_130 |
0.1043664084 |
- |
- |
PREDICTED: nifU-like protein 2, chloroplastic [Jatropha curcas] |
| 18 |
Hb_163256_020 |
0.104437034 |
- |
- |
fructokinase [Manihot esculenta] |
| 19 |
Hb_013399_050 |
0.1045849189 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 2 homolog 1-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_003768_030 |
0.1054086647 |
- |
- |
PREDICTED: ribosome biogenesis protein WDR12 homolog isoform X1 [Jatropha curcas] |