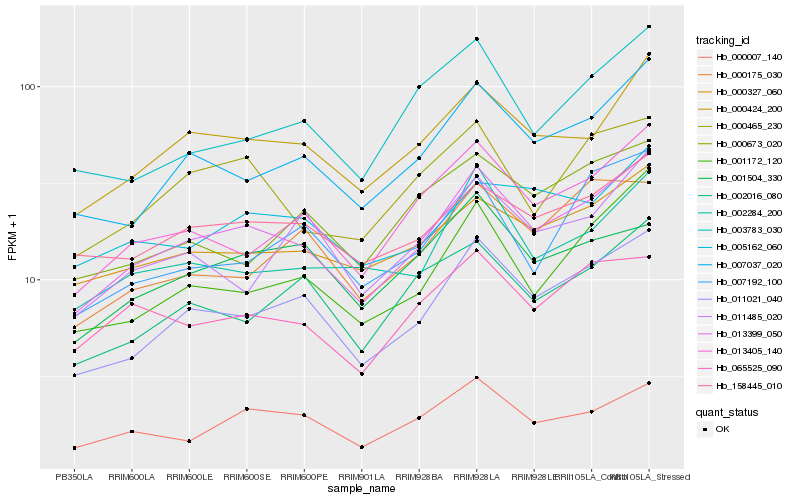
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013399_050 |
0.0 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 2 homolog 1-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_011021_040 |
0.0607552358 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 3 |
Hb_000424_200 |
0.0731127421 |
- |
- |
hypothetical protein JCGZ_07583 [Jatropha curcas] |
| 4 |
Hb_007037_020 |
0.0782910688 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 78 [Jatropha curcas] |
| 5 |
Hb_013405_140 |
0.086626438 |
- |
- |
PREDICTED: uncharacterized protein LOC105648437 [Jatropha curcas] |
| 6 |
Hb_000327_060 |
0.0875421101 |
- |
- |
PREDICTED: uncharacterized protein LOC105635258 isoform X2 [Jatropha curcas] |
| 7 |
Hb_158445_010 |
0.0915709358 |
- |
- |
PREDICTED: cysteine protease ATG4-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000007_140 |
0.0929609674 |
- |
- |
PREDICTED: F-box/FBD/LRR-repeat protein At1g13570 [Jatropha curcas] |
| 9 |
Hb_001172_120 |
0.0953900189 |
- |
- |
hypothetical protein B456_012G082000 [Gossypium raimondii] |
| 10 |
Hb_000465_230 |
0.0968283097 |
- |
- |
PREDICTED: transcriptional regulator ATRX homolog [Jatropha curcas] |
| 11 |
Hb_002284_200 |
0.0988401618 |
- |
- |
PREDICTED: probable protein phosphatase 2C 27 isoform X1 [Jatropha curcas] |
| 12 |
Hb_065525_090 |
0.0999064397 |
- |
- |
PREDICTED: CASP-like protein 4C1 [Vitis vinifera] |
| 13 |
Hb_002016_080 |
0.1015669566 |
- |
- |
PREDICTED: thiamine pyrophosphokinase 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005162_060 |
0.1045849189 |
- |
- |
PREDICTED: putative septum site-determining protein minD homolog, chloroplastic [Jatropha curcas] |
| 15 |
Hb_003783_030 |
0.106237082 |
- |
- |
hypothetical protein POPTR_0018s13980g [Populus trichocarpa] |
| 16 |
Hb_011485_020 |
0.1124163009 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_001504_330 |
0.1129576216 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g06240-like [Jatropha curcas] |
| 18 |
Hb_007192_100 |
0.1133313343 |
- |
- |
PREDICTED: protein DEHYDRATION-INDUCED 19 homolog 6-like [Jatropha curcas] |
| 19 |
Hb_000175_030 |
0.1137796385 |
- |
- |
Signal recognition particle, SRP54 subunit protein [Theobroma cacao] |
| 20 |
Hb_000673_020 |
0.1152912098 |
- |
- |
PREDICTED: ATP synthase subunit delta', mitochondrial [Jatropha curcas] |