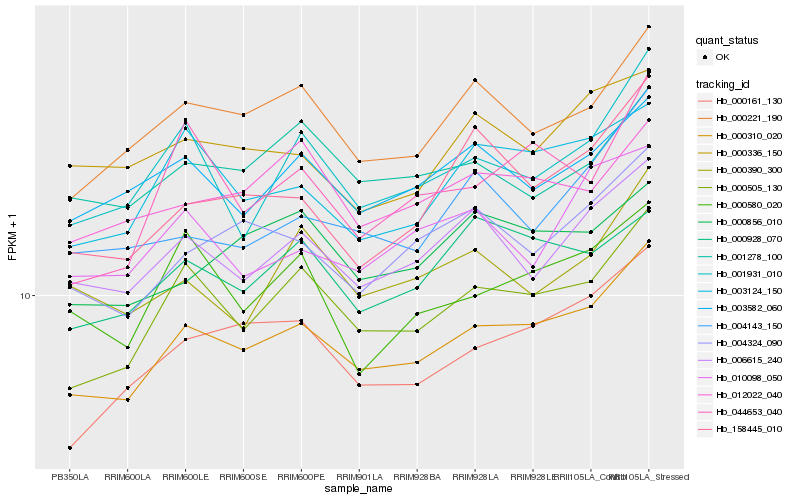
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000310_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_20797 [Jatropha curcas] |
| 2 |
Hb_000505_130 |
0.0450225731 |
- |
- |
PREDICTED: uncharacterized protein LOC105641262 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000221_190 |
0.0603709571 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 4 |
Hb_000161_130 |
0.062211096 |
- |
- |
PREDICTED: nifU-like protein 2, chloroplastic [Jatropha curcas] |
| 5 |
Hb_006615_240 |
0.0629776848 |
- |
- |
PREDICTED: uncharacterized protein LOC105640673 [Jatropha curcas] |
| 6 |
Hb_004324_090 |
0.0631986528 |
- |
- |
PREDICTED: uncharacterized protein LOC105648352 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003124_150 |
0.0663297266 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 28-like [Jatropha curcas] |
| 8 |
Hb_001931_010 |
0.0695980409 |
- |
- |
PREDICTED: NADH--cytochrome b5 reductase 1-like [Jatropha curcas] |
| 9 |
Hb_003582_060 |
0.0698972269 |
transcription factor |
TF Family: BBR-BPC |
PREDICTED: protein BASIC PENTACYSTEINE2-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_158445_010 |
0.0719336216 |
- |
- |
PREDICTED: cysteine protease ATG4-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000928_070 |
0.0750479194 |
- |
- |
PREDICTED: maltose excess protein 1, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_004143_150 |
0.0755864843 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 24-like isoform X6 [Jatropha curcas] |
| 13 |
Hb_000856_010 |
0.0758035906 |
- |
- |
PREDICTED: uncharacterized protein LOC105640466 [Jatropha curcas] |
| 14 |
Hb_012022_040 |
0.0761909065 |
- |
- |
Protein SIS1, putative [Ricinus communis] |
| 15 |
Hb_044653_040 |
0.0766377261 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase 11 [Jatropha curcas] |
| 16 |
Hb_000580_020 |
0.0778207337 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 17 |
Hb_001278_100 |
0.0778848262 |
- |
- |
PREDICTED: probable phosphopantothenoylcysteine decarboxylase [Jatropha curcas] |
| 18 |
Hb_000336_150 |
0.0798319148 |
- |
- |
ribulose-5-phosphate-3-epimerase, putative [Ricinus communis] |
| 19 |
Hb_000390_300 |
0.0805102885 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase MCC1 [Jatropha curcas] |
| 20 |
Hb_010098_050 |
0.0806427613 |
- |
- |
PREDICTED: putative deoxyribonuclease tatdn3-A [Jatropha curcas] |