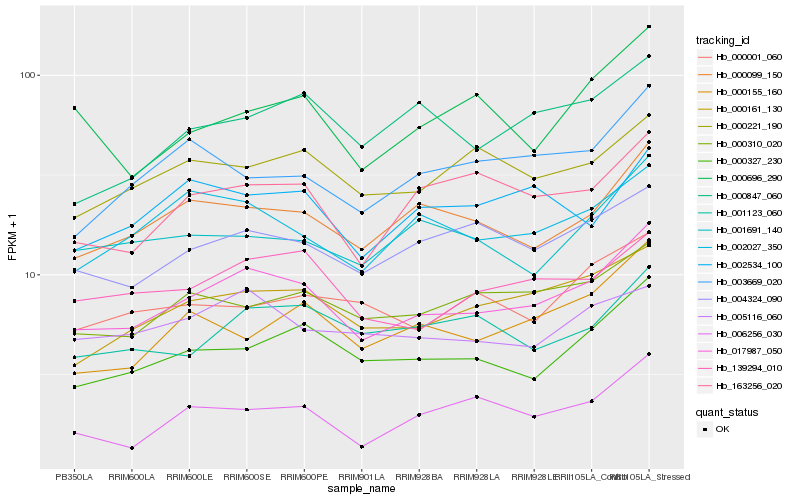
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017987_050 |
0.0 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 5, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_000161_130 |
0.0817279976 |
- |
- |
PREDICTED: nifU-like protein 2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000099_150 |
0.0859045017 |
- |
- |
PREDICTED: calcium-dependent protein kinase 3 [Jatropha curcas] |
| 4 |
Hb_000327_230 |
0.0861384058 |
- |
- |
PREDICTED: nitrilase-like protein 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_139294_010 |
0.0930831628 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_004324_090 |
0.0938932366 |
- |
- |
PREDICTED: uncharacterized protein LOC105648352 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000310_020 |
0.0955102421 |
- |
- |
hypothetical protein JCGZ_20797 [Jatropha curcas] |
| 8 |
Hb_163256_020 |
0.0976185975 |
- |
- |
fructokinase [Manihot esculenta] |
| 9 |
Hb_002027_350 |
0.0995166444 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein C-like [Jatropha curcas] |
| 10 |
Hb_001123_060 |
0.1040847524 |
- |
- |
PREDICTED: ALBINO3-like protein 2, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001691_140 |
0.1048583967 |
- |
- |
mitochondrial 50S ribosomal protein L21 [Hevea brasiliensis] |
| 12 |
Hb_000155_160 |
0.1049648131 |
- |
- |
hypothetical protein POPTR_0011s05880g [Populus trichocarpa] |
| 13 |
Hb_000221_190 |
0.1052951167 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 14 |
Hb_005116_060 |
0.1073657171 |
- |
- |
PREDICTED: mitochondrial Rho GTPase 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000847_060 |
0.1075394729 |
- |
- |
hypothetical protein [Jatropha curcas] |
| 16 |
Hb_003669_020 |
0.1087517099 |
- |
- |
PREDICTED: high mobility group nucleosome-binding domain-containing protein 5 [Populus euphratica] |
| 17 |
Hb_006256_030 |
0.1101245857 |
- |
- |
PREDICTED: putative cyclic nucleotide-gated ion channel 18 [Jatropha curcas] |
| 18 |
Hb_002534_100 |
0.1106532526 |
- |
- |
PREDICTED: malate dehydrogenase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000696_290 |
0.1106838823 |
- |
- |
PREDICTED: UPF0664 stress-induced protein C29B12.11c [Jatropha curcas] |
| 20 |
Hb_000001_060 |
0.1109492165 |
- |
- |
PREDICTED: N(6)-adenine-specific DNA methyltransferase 2 [Jatropha curcas] |