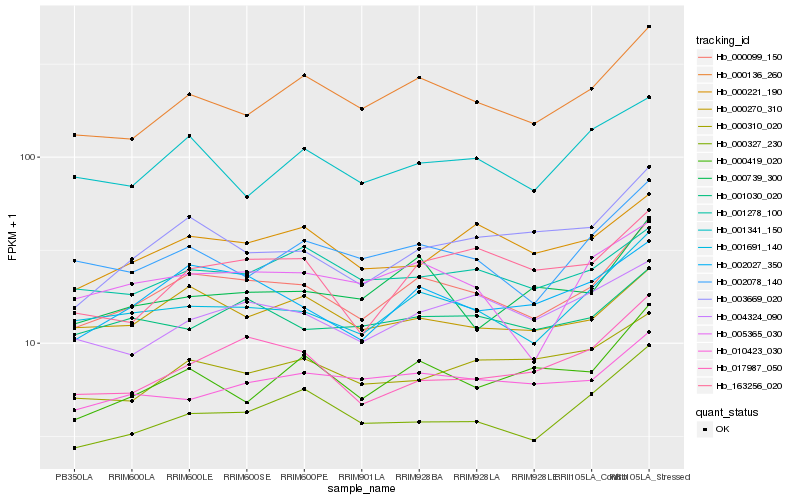
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000099_150 |
0.0 |
- |
- |
PREDICTED: calcium-dependent protein kinase 3 [Jatropha curcas] |
| 2 |
Hb_001691_140 |
0.0591455097 |
- |
- |
mitochondrial 50S ribosomal protein L21 [Hevea brasiliensis] |
| 3 |
Hb_000136_260 |
0.0706082339 |
- |
- |
40S ribosomal protein S5A [Hevea brasiliensis] |
| 4 |
Hb_002027_350 |
0.0724580015 |
- |
- |
PREDICTED: U1 small nuclear ribonucleoprotein C-like [Jatropha curcas] |
| 5 |
Hb_000327_230 |
0.0836527946 |
- |
- |
PREDICTED: nitrilase-like protein 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_017987_050 |
0.0859045017 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 5, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_002078_140 |
0.0867169294 |
- |
- |
PREDICTED: probable signal peptidase complex subunit 1 [Jatropha curcas] |
| 8 |
Hb_000270_310 |
0.087287211 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit-like [Populus euphratica] |
| 9 |
Hb_003669_020 |
0.089995054 |
- |
- |
PREDICTED: high mobility group nucleosome-binding domain-containing protein 5 [Populus euphratica] |
| 10 |
Hb_004324_090 |
0.0919958744 |
- |
- |
PREDICTED: uncharacterized protein LOC105648352 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000221_190 |
0.0921087285 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 12 |
Hb_001030_020 |
0.0930544542 |
- |
- |
Poly(A) polymerase alpha, putative [Ricinus communis] |
| 13 |
Hb_005365_030 |
0.0932625653 |
- |
- |
hypothetical protein POPTR_0002s24010g [Populus trichocarpa] |
| 14 |
Hb_001278_100 |
0.0942742744 |
- |
- |
PREDICTED: probable phosphopantothenoylcysteine decarboxylase [Jatropha curcas] |
| 15 |
Hb_000419_020 |
0.0946921113 |
- |
- |
PREDICTED: aminodeoxychorismate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_163256_020 |
0.0952020192 |
- |
- |
fructokinase [Manihot esculenta] |
| 17 |
Hb_000310_020 |
0.0963421862 |
- |
- |
hypothetical protein JCGZ_20797 [Jatropha curcas] |
| 18 |
Hb_010423_030 |
0.0964891311 |
- |
- |
PREDICTED: uncharacterized protein LOC105645584 [Jatropha curcas] |
| 19 |
Hb_001341_150 |
0.0966517018 |
- |
- |
PREDICTED: proteasome subunit beta type-4-like [Jatropha curcas] |
| 20 |
Hb_000739_300 |
0.0967383688 |
- |
- |
PREDICTED: uncharacterized protein LOC105643092 [Jatropha curcas] |