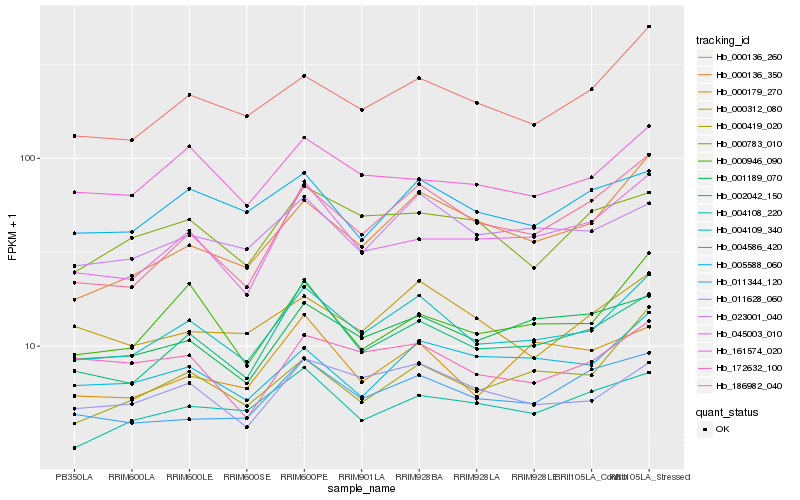
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004109_340 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14-like [Jatropha curcas] |
| 2 |
Hb_002042_150 |
0.0645536082 |
- |
- |
PREDICTED: signal peptidase complex subunit 3B [Jatropha curcas] |
| 3 |
Hb_000136_260 |
0.0730077341 |
- |
- |
40S ribosomal protein S5A [Hevea brasiliensis] |
| 4 |
Hb_011628_060 |
0.0736043005 |
- |
- |
Adenosine 3'-phospho 5'-phosphosulfate transporter, putative [Ricinus communis] |
| 5 |
Hb_023001_040 |
0.0756273921 |
- |
- |
Metallopeptidase M24 family protein isoform 1 [Theobroma cacao] |
| 6 |
Hb_161574_020 |
0.0837921064 |
- |
- |
PREDICTED: ubiquitin fusion degradation protein 1 homolog [Jatropha curcas] |
| 7 |
Hb_001189_070 |
0.0844442335 |
- |
- |
PREDICTED: ribosome production factor 1 [Jatropha curcas] |
| 8 |
Hb_005588_060 |
0.0846132905 |
- |
- |
PREDICTED: succinate dehydrogenase subunit 5, mitochondrial [Jatropha curcas] |
| 9 |
Hb_004586_420 |
0.0847038801 |
- |
- |
PREDICTED: uncharacterized protein LOC105630659 [Jatropha curcas] |
| 10 |
Hb_000312_080 |
0.0859233905 |
- |
- |
PREDICTED: transmembrane protein 97-like [Jatropha curcas] |
| 11 |
Hb_011344_120 |
0.0862705896 |
- |
- |
PREDICTED: uncharacterized protein LOC105645969 [Jatropha curcas] |
| 12 |
Hb_045003_010 |
0.0864793394 |
- |
- |
Stomatin-1, putative [Ricinus communis] |
| 13 |
Hb_186982_040 |
0.0877383037 |
- |
- |
hypothetical protein PHAVU_003G089200g [Phaseolus vulgaris] |
| 14 |
Hb_000946_090 |
0.0878021479 |
- |
- |
PREDICTED: cyclin-C1-2-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_004108_220 |
0.0880308459 |
- |
- |
PREDICTED: mRNA-capping enzyme-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000419_020 |
0.0884041303 |
- |
- |
PREDICTED: aminodeoxychorismate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_000136_350 |
0.0890995575 |
- |
- |
PREDICTED: 40S ribosomal protein S5 [Nomascus leucogenys] |
| 18 |
Hb_000783_010 |
0.0916210517 |
- |
- |
PREDICTED: ras-related protein RABF1 [Jatropha curcas] |
| 19 |
Hb_172632_100 |
0.0926858041 |
- |
- |
PREDICTED: reticulon-like protein B22 [Jatropha curcas] |
| 20 |
Hb_000179_270 |
0.0932750683 |
- |
- |
hypothetical protein JCGZ_07444 [Jatropha curcas] |