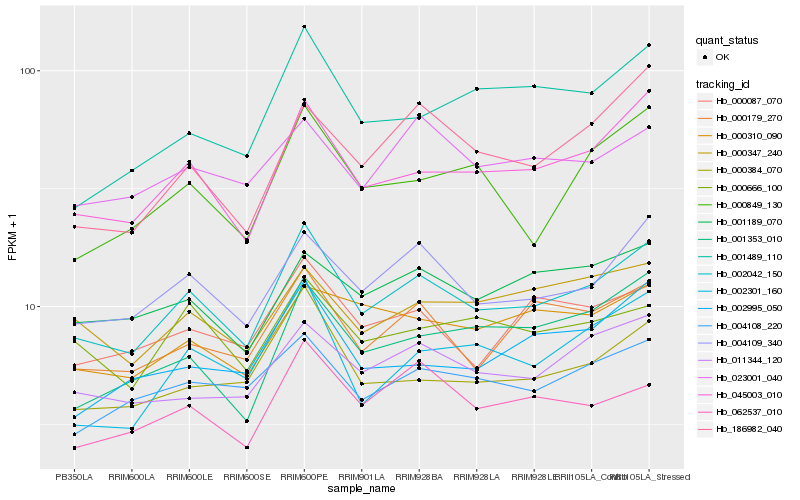
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002042_150 |
0.0 |
- |
- |
PREDICTED: signal peptidase complex subunit 3B [Jatropha curcas] |
| 2 |
Hb_045003_010 |
0.0566498986 |
- |
- |
Stomatin-1, putative [Ricinus communis] |
| 3 |
Hb_004109_340 |
0.0645536082 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 14-like [Jatropha curcas] |
| 4 |
Hb_000179_270 |
0.0781234649 |
- |
- |
hypothetical protein JCGZ_07444 [Jatropha curcas] |
| 5 |
Hb_000384_070 |
0.0797105362 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_011344_120 |
0.0824057811 |
- |
- |
PREDICTED: uncharacterized protein LOC105645969 [Jatropha curcas] |
| 7 |
Hb_004108_220 |
0.0839922848 |
- |
- |
PREDICTED: mRNA-capping enzyme-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_062537_010 |
0.0842336093 |
- |
- |
hypothetical protein JCGZ_13884 [Jatropha curcas] |
| 9 |
Hb_000087_070 |
0.0860051937 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 10 |
Hb_001189_070 |
0.0866865659 |
- |
- |
PREDICTED: ribosome production factor 1 [Jatropha curcas] |
| 11 |
Hb_002301_160 |
0.0887164284 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04780-like [Jatropha curcas] |
| 12 |
Hb_000666_100 |
0.0892889203 |
- |
- |
Actin-related protein 2/3 complex subunit 2 isoform 2 [Theobroma cacao] |
| 13 |
Hb_000849_130 |
0.0917869874 |
- |
- |
PREDICTED: adrenodoxin-like protein, mitochondrial [Nelumbo nucifera] |
| 14 |
Hb_000347_240 |
0.0919012489 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Pyrus x bretschneideri] |
| 15 |
Hb_001353_010 |
0.0923400425 |
- |
- |
PREDICTED: cytochrome c oxidase assembly protein COX11, mitochondrial [Jatropha curcas] |
| 16 |
Hb_023001_040 |
0.0927437478 |
- |
- |
Metallopeptidase M24 family protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_002995_050 |
0.0940062238 |
- |
- |
PREDICTED: carboxy-terminal domain RNA polymerase II polypeptide A small phosphatase 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000310_090 |
0.0940995733 |
- |
- |
3-oxoacyl-[acyl-carrier-protein] synthase 3 A, chloroplastic [Jatropha curcas] |
| 19 |
Hb_186982_040 |
0.094261622 |
- |
- |
hypothetical protein PHAVU_003G089200g [Phaseolus vulgaris] |
| 20 |
Hb_001489_110 |
0.0942961404 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 36 kDa-like [Citrus sinensis] |