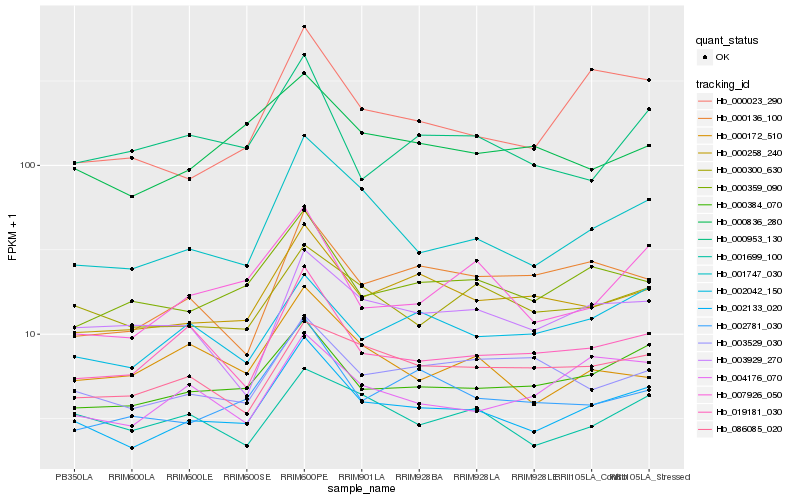
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002133_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001747_030 |
0.0879107614 |
- |
- |
hypothetical protein F383_08210 [Gossypium arboreum] |
| 3 |
Hb_000384_070 |
0.091512151 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000258_240 |
0.0994492752 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 5 |
Hb_000359_090 |
0.1070800724 |
- |
- |
PREDICTED: persulfide dioxygenase ETHE1 homolog, mitochondrial-like [Malus domestica] |
| 6 |
Hb_019181_030 |
0.1097548839 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: conserved oligomeric Golgi complex subunit 6-like [Pyrus x bretschneideri] |
| 7 |
Hb_002781_030 |
0.1129117977 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 8 |
Hb_007926_050 |
0.1145616492 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 9 |
Hb_000300_630 |
0.1148903781 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 7 [Jatropha curcas] |
| 10 |
Hb_000023_290 |
0.116184277 |
- |
- |
steroid binding protein, putative [Ricinus communis] |
| 11 |
Hb_002042_150 |
0.1200477708 |
- |
- |
PREDICTED: signal peptidase complex subunit 3B [Jatropha curcas] |
| 12 |
Hb_000836_280 |
0.1202477488 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001699_100 |
0.1213175805 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 14 |
Hb_004176_070 |
0.1236950333 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_003929_270 |
0.1250985165 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000953_130 |
0.1270407668 |
- |
- |
PREDICTED: fructose-bisphosphate aldolase cytoplasmic isozyme [Jatropha curcas] |
| 17 |
Hb_086085_020 |
0.1279088462 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein isoform 3 [Theobroma cacao] |
| 18 |
Hb_003529_030 |
0.1282886202 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein, putative [Ricinus communis] |
| 19 |
Hb_000136_100 |
0.1309314341 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 20 |
Hb_000172_510 |
0.1312604783 |
- |
- |
hypothetical protein B456_007G243100 [Gossypium raimondii] |