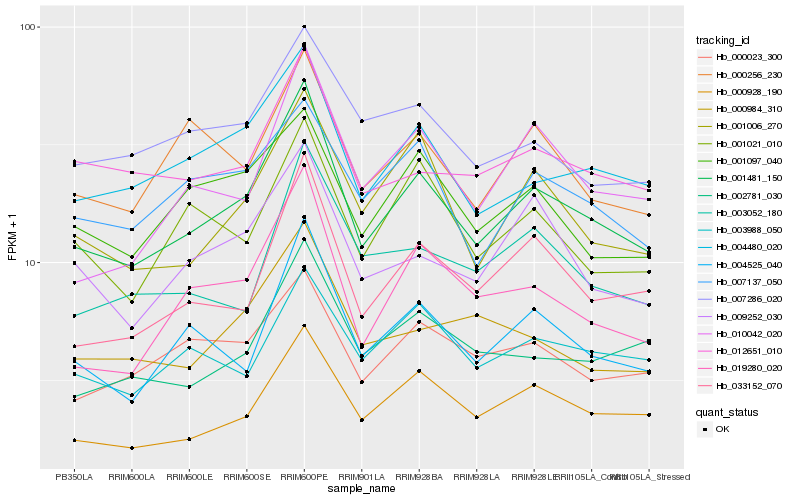
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001481_150 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s15440g [Populus trichocarpa] |
| 2 |
Hb_004480_020 |
0.0863456751 |
- |
- |
PREDICTED: uncharacterized protein LOC105637166 [Jatropha curcas] |
| 3 |
Hb_000928_190 |
0.0874099902 |
- |
- |
PREDICTED: putative acyl-activating enzyme 19 isoform X1 [Jatropha curcas] |
| 4 |
Hb_033152_070 |
0.087867864 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 5 |
Hb_004525_040 |
0.08897559 |
- |
- |
PREDICTED: sugar transporter ERD6-like 6 [Jatropha curcas] |
| 6 |
Hb_019280_020 |
0.0894064276 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK14 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001006_270 |
0.0967523503 |
- |
- |
peptide transporter, putative [Ricinus communis] |
| 8 |
Hb_010042_020 |
0.1009542175 |
- |
- |
UDP-n-acteylglucosamine pyrophosphorylase, putative [Ricinus communis] |
| 9 |
Hb_009252_030 |
0.102308167 |
- |
- |
PREDICTED: CASP-like protein 4A3 [Jatropha curcas] |
| 10 |
Hb_002781_030 |
0.1031191379 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_012651_010 |
0.1056972047 |
- |
- |
PREDICTED: uncharacterized protein LOC105645131 [Jatropha curcas] |
| 12 |
Hb_000023_300 |
0.1071655172 |
- |
- |
Ubiquitin carboxyl-terminal hydrolase, putative [Ricinus communis] |
| 13 |
Hb_001097_040 |
0.1081202782 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 14 |
Hb_001021_010 |
0.108335048 |
- |
- |
PREDICTED: bifunctional protein FolD 2 [Jatropha curcas] |
| 15 |
Hb_000256_230 |
0.108369793 |
- |
- |
PREDICTED: uncharacterized protein LOC105637594 [Jatropha curcas] |
| 16 |
Hb_007137_050 |
0.1083917225 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_003052_180 |
0.1100127141 |
- |
- |
Diacylglycerol Cholinephosphotransferase [Ricinus communis] |
| 18 |
Hb_000984_310 |
0.1101169168 |
- |
- |
Hydrolase, hydrolyzing O-glycosyl compounds, putative [Theobroma cacao] |
| 19 |
Hb_007286_020 |
0.1103548963 |
- |
- |
hypothetical protein L484_010675 [Morus notabilis] |
| 20 |
Hb_003988_050 |
0.1116490081 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |