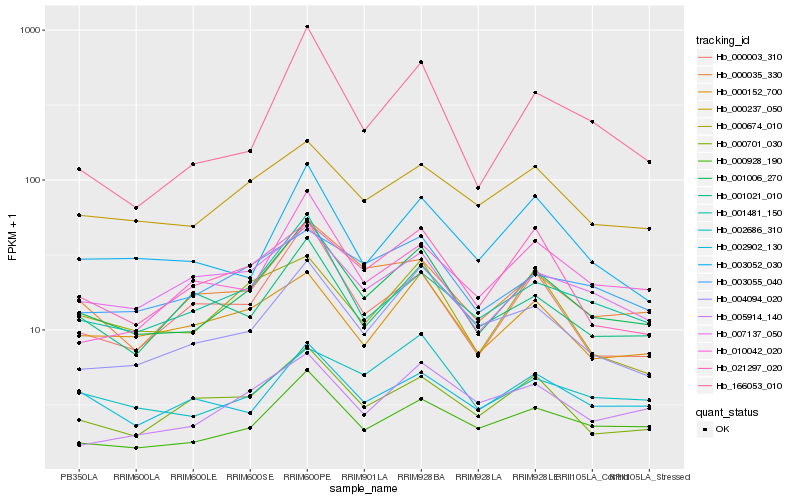
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001006_270 |
0.0 |
- |
- |
peptide transporter, putative [Ricinus communis] |
| 2 |
Hb_000928_190 |
0.0915506725 |
- |
- |
PREDICTED: putative acyl-activating enzyme 19 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001481_150 |
0.0967523503 |
- |
- |
hypothetical protein POPTR_0001s15440g [Populus trichocarpa] |
| 4 |
Hb_000035_330 |
0.1005731887 |
- |
- |
Transmembrane protein 85 [Theobroma cacao] |
| 5 |
Hb_000237_050 |
0.1018532307 |
- |
- |
CP2 [Hevea brasiliensis] |
| 6 |
Hb_004094_020 |
0.1071573233 |
- |
- |
Pyruvate kinase family protein isoform 1 [Theobroma cacao] |
| 7 |
Hb_000701_030 |
0.1075503643 |
- |
- |
Sodium/hydrogen exchanger 6 -like protein [Gossypium arboreum] |
| 8 |
Hb_000003_310 |
0.1075691667 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 9 |
Hb_003055_040 |
0.112458637 |
- |
- |
PREDICTED: secretory carrier-associated membrane protein 3-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000152_700 |
0.1135601695 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIN2 [Jatropha curcas] |
| 11 |
Hb_007137_050 |
0.1147428317 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 12 |
Hb_001021_010 |
0.115772502 |
- |
- |
PREDICTED: bifunctional protein FolD 2 [Jatropha curcas] |
| 13 |
Hb_003052_030 |
0.11758177 |
- |
- |
6-phosphogluconate dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_021297_020 |
0.1183957018 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 15 |
Hb_166053_010 |
0.1195432976 |
- |
- |
glyceraldehyde-3-phosphate dehydrogenase, partial [Vernicia fordii] |
| 16 |
Hb_002902_130 |
0.1196390215 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 17 |
Hb_005914_140 |
0.1210527258 |
- |
- |
PREDICTED: uncharacterized protein LOC105632183 [Jatropha curcas] |
| 18 |
Hb_002686_310 |
0.1215055539 |
- |
- |
PREDICTED: aquaporin SIP1-1 [Jatropha curcas] |
| 19 |
Hb_000674_010 |
0.1216142753 |
- |
- |
PREDICTED: copper-transporting ATPase RAN1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_010042_020 |
0.1221640889 |
- |
- |
UDP-n-acteylglucosamine pyrophosphorylase, putative [Ricinus communis] |