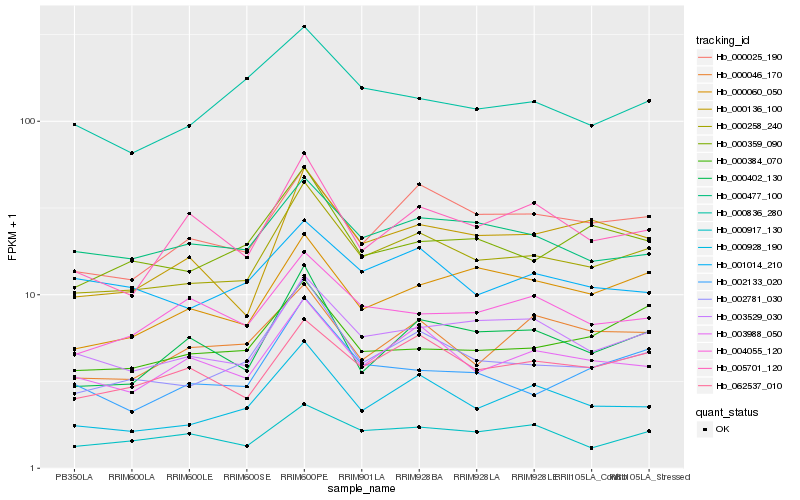
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000258_240 |
0.0 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 2 |
Hb_002781_030 |
0.0603115989 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_003529_030 |
0.0722838705 |
- |
- |
Non-imprinted in Prader-Willi/Angelman syndrome region protein, putative [Ricinus communis] |
| 4 |
Hb_003988_050 |
0.0782848373 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 5 |
Hb_062537_010 |
0.0810967329 |
- |
- |
hypothetical protein JCGZ_13884 [Jatropha curcas] |
| 6 |
Hb_000928_190 |
0.0833530637 |
- |
- |
PREDICTED: putative acyl-activating enzyme 19 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000025_190 |
0.0837588815 |
- |
- |
26S proteasome non-atpase regulatory subunit, putative [Ricinus communis] |
| 8 |
Hb_000477_100 |
0.0891916548 |
- |
- |
PREDICTED: GDP-mannose transporter GONST4 [Jatropha curcas] |
| 9 |
Hb_000359_090 |
0.0899279999 |
- |
- |
PREDICTED: persulfide dioxygenase ETHE1 homolog, mitochondrial-like [Malus domestica] |
| 10 |
Hb_000917_130 |
0.0934232578 |
- |
- |
PREDICTED: uncharacterized protein LOC105635730 [Jatropha curcas] |
| 11 |
Hb_000402_130 |
0.0946134766 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 12 |
Hb_000060_050 |
0.0970932383 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 13 |
Hb_000136_100 |
0.0978491318 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 14 |
Hb_001014_210 |
0.0993518507 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 15 |
Hb_002133_020 |
0.0994492752 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000836_280 |
0.1000328376 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_005701_120 |
0.1012057496 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 18 |
Hb_000384_070 |
0.1020856572 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000046_170 |
0.1032356466 |
- |
- |
PREDICTED: uncharacterized protein LOC105631841 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004055_120 |
0.1032566179 |
- |
- |
PREDICTED: uncharacterized protein LOC105631443 [Jatropha curcas] |