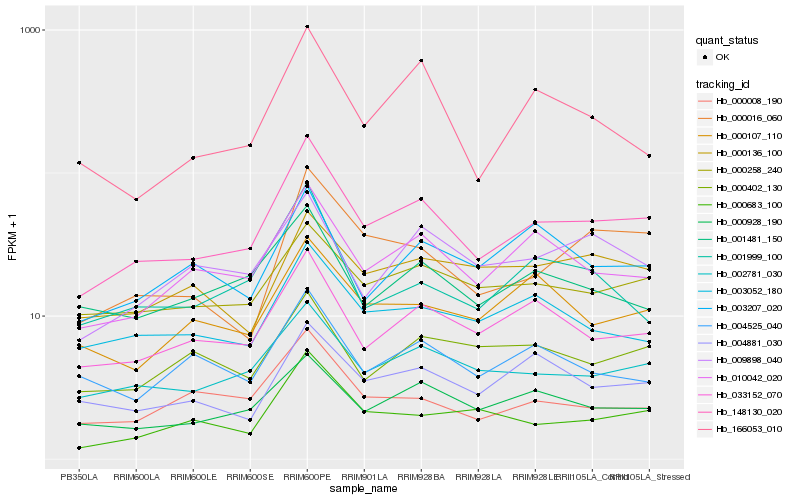
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_148130_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_002781_030 |
0.0989908202 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_010042_020 |
0.108591951 |
- |
- |
UDP-n-acteylglucosamine pyrophosphorylase, putative [Ricinus communis] |
| 4 |
Hb_033152_070 |
0.1094981191 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 5 |
Hb_000928_190 |
0.1160480289 |
- |
- |
PREDICTED: putative acyl-activating enzyme 19 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000258_240 |
0.1237156315 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 7 |
Hb_000008_190 |
0.1268626416 |
- |
- |
PREDICTED: protein ENHANCED DISEASE RESISTANCE 2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000016_060 |
0.12856045 |
- |
- |
mads box protein, putative [Ricinus communis] |
| 9 |
Hb_003052_180 |
0.128866686 |
- |
- |
Diacylglycerol Cholinephosphotransferase [Ricinus communis] |
| 10 |
Hb_001481_150 |
0.1297223447 |
- |
- |
hypothetical protein POPTR_0001s15440g [Populus trichocarpa] |
| 11 |
Hb_000683_100 |
0.1304655624 |
- |
- |
hypothetical protein JCGZ_22137 [Jatropha curcas] |
| 12 |
Hb_004881_030 |
0.1355357821 |
- |
- |
PREDICTED: uncharacterized protein LOC105629532 isoform X1 [Jatropha curcas] |
| 13 |
Hb_009898_040 |
0.1370131588 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000107_110 |
0.1416193325 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 15 |
Hb_004525_040 |
0.1425421612 |
- |
- |
PREDICTED: sugar transporter ERD6-like 6 [Jatropha curcas] |
| 16 |
Hb_001999_100 |
0.1426845273 |
- |
- |
PREDICTED: UDP-glycosyltransferase 71D1-like [Jatropha curcas] |
| 17 |
Hb_003207_020 |
0.144286256 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 18 |
Hb_166053_010 |
0.1445140995 |
- |
- |
glyceraldehyde-3-phosphate dehydrogenase, partial [Vernicia fordii] |
| 19 |
Hb_000402_130 |
0.1445438861 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 20 |
Hb_000136_100 |
0.1458684453 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |