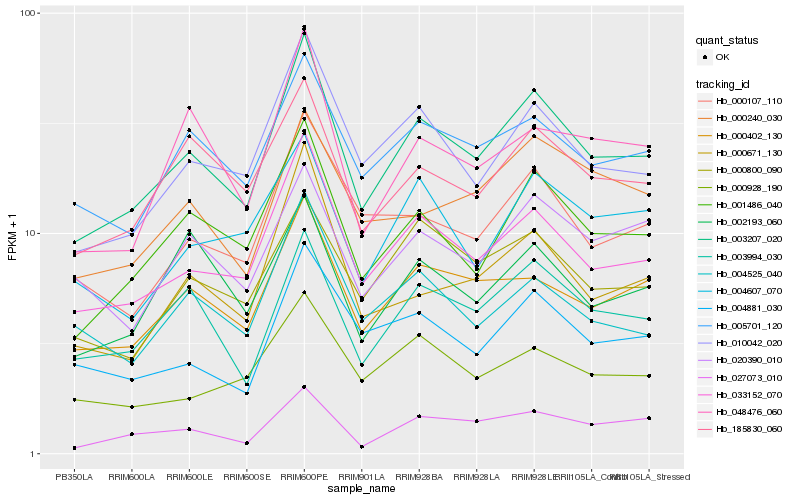
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003207_020 |
0.0 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 2 |
Hb_033152_070 |
0.061326993 |
- |
- |
N-acetyl-gamma-glutamyl-phosphate reductase, putative [Ricinus communis] |
| 3 |
Hb_001486_040 |
0.0736077659 |
- |
- |
PREDICTED: uncharacterized protein LOC105632624 isoform X1 [Jatropha curcas] |
| 4 |
Hb_010042_020 |
0.0822482597 |
- |
- |
UDP-n-acteylglucosamine pyrophosphorylase, putative [Ricinus communis] |
| 5 |
Hb_003994_030 |
0.0857079854 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 6 |
Hb_000402_130 |
0.0877831655 |
- |
- |
PREDICTED: AP-1 complex subunit mu-2 [Jatropha curcas] |
| 7 |
Hb_005701_120 |
0.0930038638 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 8 |
Hb_185830_060 |
0.095725762 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 9 |
Hb_000107_110 |
0.105540291 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 10 |
Hb_004881_030 |
0.1059072687 |
- |
- |
PREDICTED: uncharacterized protein LOC105629532 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000800_090 |
0.1063997834 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 12 |
Hb_027073_010 |
0.1084432014 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 13 |
Hb_048476_060 |
0.1085572089 |
- |
- |
PREDICTED: CRAL-TRIO domain-containing protein YKL091C-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_004525_040 |
0.1089815548 |
- |
- |
PREDICTED: sugar transporter ERD6-like 6 [Jatropha curcas] |
| 15 |
Hb_000671_130 |
0.1109630394 |
- |
- |
PREDICTED: CMP-sialic acid transporter 2 isoform X2 [Jatropha curcas] |
| 16 |
Hb_020390_010 |
0.111109163 |
- |
- |
PREDICTED: expansin-A13 [Jatropha curcas] |
| 17 |
Hb_004607_070 |
0.1114594047 |
- |
- |
integral membrane protein, putative [Ricinus communis] |
| 18 |
Hb_000928_190 |
0.1123744306 |
- |
- |
PREDICTED: putative acyl-activating enzyme 19 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002193_060 |
0.1123917379 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 20 |
Hb_000240_030 |
0.1129879705 |
- |
- |
conserved hypothetical protein [Ricinus communis] |