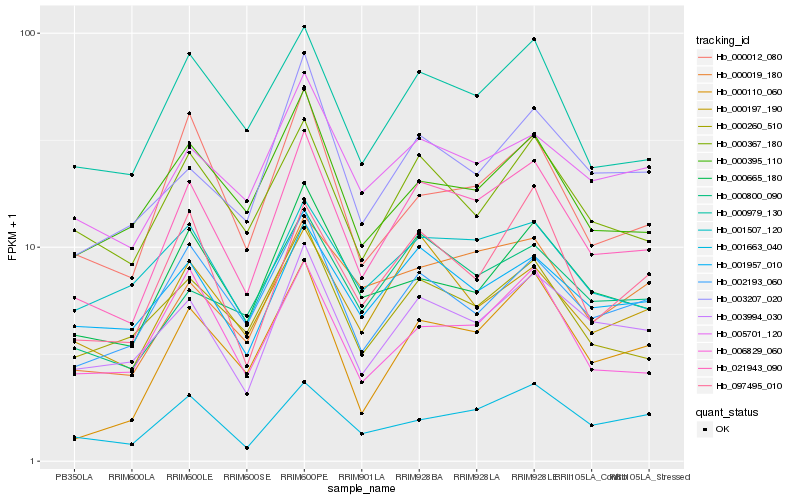
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021943_090 |
0.0 |
- |
- |
PREDICTED: probable methylenetetrahydrofolate reductase [Jatropha curcas] |
| 2 |
Hb_003994_030 |
0.0898244623 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 3 |
Hb_000979_130 |
0.0906705119 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 4 |
Hb_000367_180 |
0.0998215697 |
- |
- |
Heparanase-2, putative [Ricinus communis] |
| 5 |
Hb_000260_510 |
0.1027890129 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 6 |
Hb_000800_090 |
0.1049354361 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 1 of pyruvate dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000665_180 |
0.1055469354 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 8 |
Hb_002193_060 |
0.1056385421 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 9 |
Hb_000197_190 |
0.1056934041 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |
| 10 |
Hb_001957_010 |
0.106643052 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g14410 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001663_040 |
0.1073282251 |
- |
- |
hypothetical protein B456_007G162000 [Gossypium raimondii] |
| 12 |
Hb_005701_120 |
0.1077069104 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Cucumis sativus] |
| 13 |
Hb_097495_010 |
0.1091986076 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X2 [Jatropha curcas] |
| 14 |
Hb_000012_080 |
0.1111664145 |
- |
- |
PREDICTED: clathrin interactor EPSIN 1 [Jatropha curcas] |
| 15 |
Hb_000395_110 |
0.1113246798 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 16 |
Hb_006829_060 |
0.1126768086 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000110_060 |
0.1127706806 |
- |
- |
PREDICTED: uncharacterized protein LOC105636258 [Jatropha curcas] |
| 18 |
Hb_003207_020 |
0.1138320904 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 19 |
Hb_000019_180 |
0.1153174521 |
transcription factor |
TF Family: SET |
Histone-lysine N-methyltransferase family protein [Populus trichocarpa] |
| 20 |
Hb_001507_120 |
0.1162713313 |
- |
- |
transcription factor, putative [Ricinus communis] |