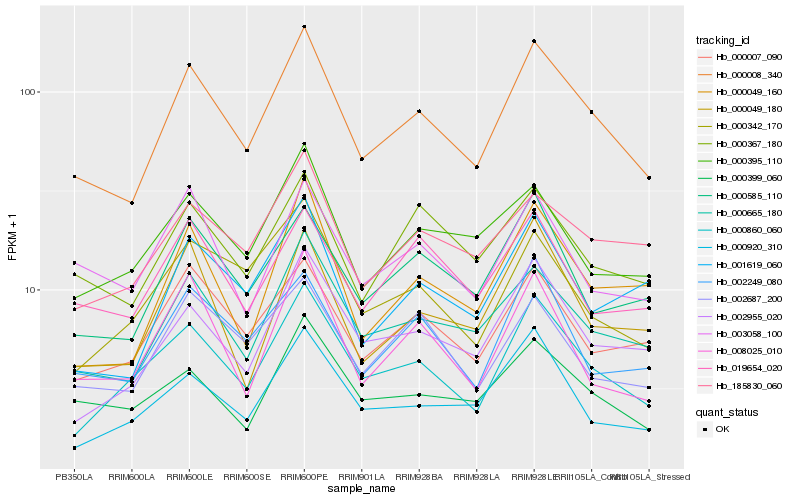
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000008_340 |
0.0 |
- |
- |
PREDICTED: aquaporin PIP2-4 [Jatropha curcas] |
| 2 |
Hb_000665_180 |
0.0765883462 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 3 |
Hb_000860_060 |
0.0921682742 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 [Jatropha curcas] |
| 4 |
Hb_000585_110 |
0.0996235866 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 5 |
Hb_000007_090 |
0.0999087204 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 6 |
Hb_002955_020 |
0.1003845722 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 7 |
Hb_000049_160 |
0.1023271168 |
- |
- |
tyrosine decarboxylase family protein [Populus trichocarpa] |
| 8 |
Hb_002249_080 |
0.1035839634 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 9 |
Hb_000367_180 |
0.1045619854 |
- |
- |
Heparanase-2, putative [Ricinus communis] |
| 10 |
Hb_185830_060 |
0.1076629606 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 11 |
Hb_001619_060 |
0.10902171 |
- |
- |
PREDICTED: uncharacterized methyltransferase At1g78140, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000399_060 |
0.1100490162 |
- |
- |
PREDICTED: uncharacterized protein LOC105649658 [Jatropha curcas] |
| 13 |
Hb_002687_200 |
0.1101306528 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000920_310 |
0.1126772788 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 15 |
Hb_000049_180 |
0.1128415675 |
- |
- |
aromatic amino acid decarboxylase, putative [Ricinus communis] |
| 16 |
Hb_000342_170 |
0.1131973917 |
- |
- |
PREDICTED: uncharacterized protein LOC100807540 isoform X2 [Glycine max] |
| 17 |
Hb_000395_110 |
0.1133960855 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 18 |
Hb_008025_010 |
0.1141000153 |
- |
- |
PREDICTED: lysosomal beta glucosidase-like [Musa acuminata subsp. malaccensis] |
| 19 |
Hb_003058_100 |
0.1142351509 |
- |
- |
PREDICTED: hydroxymethylglutaryl-CoA lyase, mitochondrial [Jatropha curcas] |
| 20 |
Hb_019654_020 |
0.1142835664 |
- |
- |
PREDICTED: T-complex protein 1 subunit beta [Jatropha curcas] |