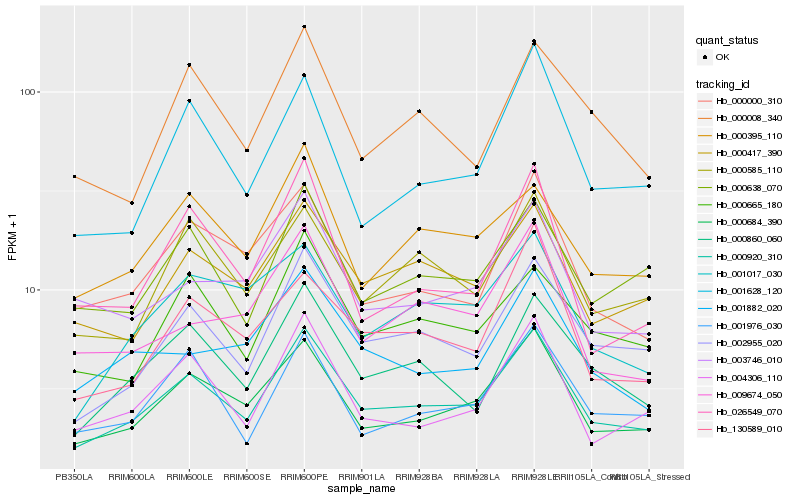
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000920_310 |
0.0 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 2 |
Hb_000684_390 |
0.0672467117 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 3 |
Hb_002955_020 |
0.0741278731 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_000860_060 |
0.0920551793 |
transcription factor |
TF Family: SBP |
PREDICTED: squamosa promoter-binding-like protein 6 [Jatropha curcas] |
| 5 |
Hb_000417_390 |
0.0960709334 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor A [Jatropha curcas] |
| 6 |
Hb_004306_110 |
0.1012644201 |
transcription factor |
TF Family: Trihelix |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH9 [Jatropha curcas] |
| 7 |
Hb_000000_310 |
0.1020911641 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 8 |
Hb_000665_180 |
0.1038686845 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 9 |
Hb_000638_070 |
0.1044618571 |
- |
- |
PREDICTED: protein trichome birefringence-like 14 [Jatropha curcas] |
| 10 |
Hb_000585_110 |
0.1047215441 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 11 |
Hb_001976_030 |
0.1051102185 |
- |
- |
PREDICTED: protein NETWORKED 2D-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_001628_120 |
0.1053011035 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |
| 13 |
Hb_130589_010 |
0.1060617828 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 14 |
Hb_000395_110 |
0.1063546854 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 15 |
Hb_003746_010 |
0.1110504067 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001017_030 |
0.1114299277 |
- |
- |
copine, putative [Ricinus communis] |
| 17 |
Hb_001882_020 |
0.1117404214 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 18 |
Hb_000008_340 |
0.1126772788 |
- |
- |
PREDICTED: aquaporin PIP2-4 [Jatropha curcas] |
| 19 |
Hb_009674_050 |
0.1134092541 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 20 |
Hb_026549_070 |
0.1141536371 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |