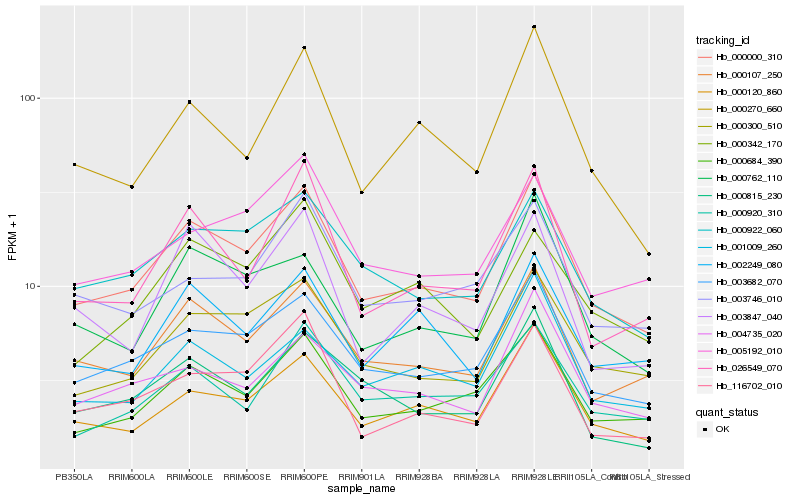
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000000_310 |
0.0 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 2 |
Hb_003682_070 |
0.0618714137 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000107_250 |
0.0717016784 |
- |
- |
voltage-gated clc-type chloride channel, putative [Ricinus communis] |
| 4 |
Hb_000300_510 |
0.0773687224 |
- |
- |
PREDICTED: uncharacterized protein LOC105632605 [Jatropha curcas] |
| 5 |
Hb_000684_390 |
0.0791818362 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 6 |
Hb_000120_860 |
0.0801553845 |
- |
- |
nucellin, putative [Ricinus communis] |
| 7 |
Hb_000922_060 |
0.0832653569 |
- |
- |
kinesin light chain, putative [Ricinus communis] |
| 8 |
Hb_026549_070 |
0.097362583 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 9 |
Hb_000270_660 |
0.1008210233 |
- |
- |
purine permease, putative [Ricinus communis] |
| 10 |
Hb_000920_310 |
0.1020911641 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 11 |
Hb_002249_080 |
0.1026014745 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 12 |
Hb_116702_010 |
0.1027703002 |
- |
- |
Leucine-rich repeat protein kinase family protein [Theobroma cacao] |
| 13 |
Hb_005192_010 |
0.1040572492 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 14 |
Hb_000762_110 |
0.1046417499 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000815_230 |
0.1056636706 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_004735_020 |
0.1059883819 |
- |
- |
PREDICTED: uncharacterized protein LOC105639315 [Jatropha curcas] |
| 17 |
Hb_003746_010 |
0.1061622764 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003847_040 |
0.1094087253 |
- |
- |
adenosine diphosphatase, putative [Ricinus communis] |
| 19 |
Hb_001009_260 |
0.1108888061 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000342_170 |
0.1112119291 |
- |
- |
PREDICTED: uncharacterized protein LOC100807540 isoform X2 [Glycine max] |