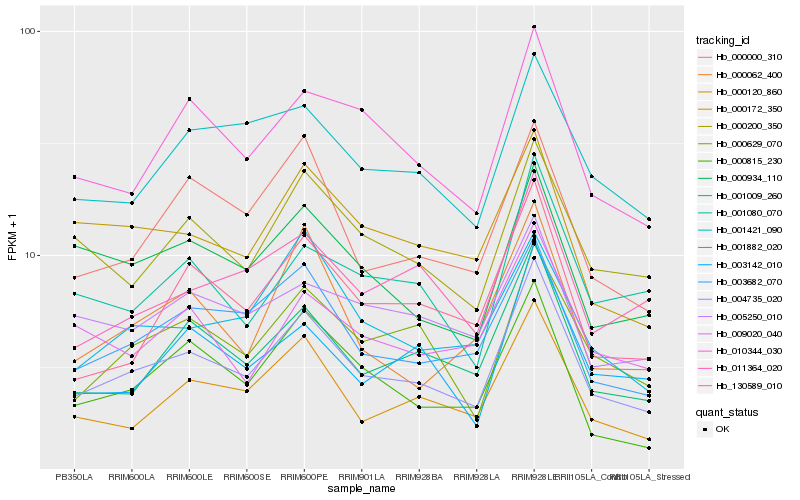
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004735_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639315 [Jatropha curcas] |
| 2 |
Hb_001009_260 |
0.0861485537 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000120_860 |
0.0900264986 |
- |
- |
nucellin, putative [Ricinus communis] |
| 4 |
Hb_003682_070 |
0.0902145493 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000629_070 |
0.0980523268 |
- |
- |
DNA-3-methyladenine glycosylase, putative [Ricinus communis] |
| 6 |
Hb_010344_030 |
0.0998638736 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_130589_010 |
0.1044361868 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 8 |
Hb_000200_350 |
0.105794812 |
- |
- |
PREDICTED: uncharacterized protein LOC105636929 [Jatropha curcas] |
| 9 |
Hb_000000_310 |
0.1059883819 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 10 |
Hb_000815_230 |
0.1077707259 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001421_090 |
0.1097150524 |
- |
- |
hypothetical protein RCOM_1278080 [Ricinus communis] |
| 12 |
Hb_003142_010 |
0.1114332969 |
transcription factor |
TF Family: SET |
PREDICTED: LOW QUALITY PROTEIN: histone-lysine N-methyltransferase ATX5-like [Populus euphratica] |
| 13 |
Hb_000172_350 |
0.1121150827 |
- |
- |
PREDICTED: uncharacterized protein LOC105650963 [Jatropha curcas] |
| 14 |
Hb_000934_110 |
0.1125269546 |
- |
- |
PREDICTED: protein trichome berefringence-like 7 [Jatropha curcas] |
| 15 |
Hb_005250_010 |
0.1127169443 |
- |
- |
hypothetical protein JCGZ_15339 [Jatropha curcas] |
| 16 |
Hb_011364_020 |
0.1131405833 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000062_400 |
0.1133456134 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001882_020 |
0.1147344011 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 19 |
Hb_001080_070 |
0.1149082138 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 20 |
Hb_009020_040 |
0.1150749118 |
- |
- |
PREDICTED: triacylglycerol lipase 1 isoform X1 [Jatropha curcas] |