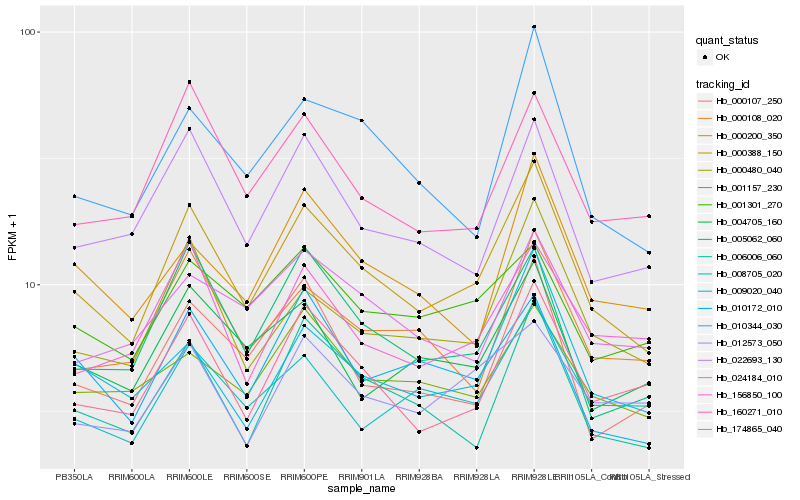
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000388_150 |
0.0 |
- |
- |
hypothetical protein JCGZ_21651 [Jatropha curcas] |
| 2 |
Hb_000480_040 |
0.0890213437 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 3 |
Hb_006006_060 |
0.0905439327 |
- |
- |
PREDICTED: triacylglycerol lipase 1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_174865_040 |
0.0920813581 |
- |
- |
PREDICTED: crt homolog 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_010344_030 |
0.0976533556 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_022693_130 |
0.0987706174 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_005062_060 |
0.0992435319 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 8 |
Hb_009020_040 |
0.1001372299 |
- |
- |
PREDICTED: triacylglycerol lipase 1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_010172_010 |
0.1036350566 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 20 [Jatropha curcas] |
| 10 |
Hb_000107_250 |
0.1048322845 |
- |
- |
voltage-gated clc-type chloride channel, putative [Ricinus communis] |
| 11 |
Hb_160271_010 |
0.1055028564 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_000200_350 |
0.1066687014 |
- |
- |
PREDICTED: uncharacterized protein LOC105636929 [Jatropha curcas] |
| 13 |
Hb_000108_020 |
0.107040217 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 14 |
Hb_004705_160 |
0.1099951288 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 15 |
Hb_001157_230 |
0.1109965327 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 16 |
Hb_008705_020 |
0.1117407648 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 17 |
Hb_012573_050 |
0.1119338753 |
- |
- |
choline/ethanolamine kinase family protein [Populus trichocarpa] |
| 18 |
Hb_001301_270 |
0.1120123899 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 19 |
Hb_156850_100 |
0.1123610575 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 20 |
Hb_024184_010 |
0.1133490355 |
- |
- |
PREDICTED: clustered mitochondria protein-like isoform X1 [Citrus sinensis] |